Topic #5: Protein Function #3--Receptors & Cellular Signaling
1/48
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
49 Terms
Cellular Signaling
def:Production, release, detection, & response to extracellular signaling (primary way cell comm.) molecules is key regulatory & control mechanism in human body
A. They carry information that controls numerous biochemical & physiological processes (e.g., gene expression, metabolism, & growth & differentiation)
B. Large variety of molecules are utilized (e.g., small organic molecules,
lipids, peptides, proteins, etc.)
II. Signaling cells (produce & secrete extracellular signaling molecule, i.e., ligand) → Target cells (respond to ligand via receptor)
Juxtacrine(Type of cellular signaling)
cell produces signalling molecule bound in its membrane, target cell has receptor for signalling molecule in its membrane ( 2 cells must get close to eachother so signal and receptor can bond)-important in immune cells
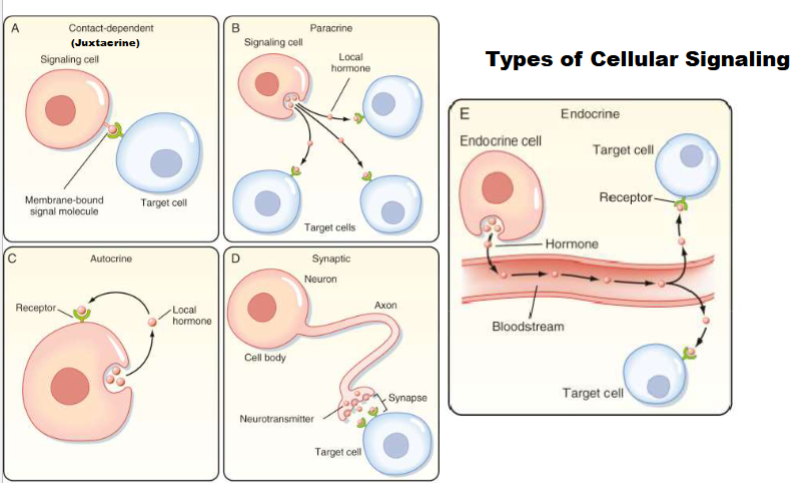
Paracrine (Type of cellular signaling)
cell and target cell in general area of each other, release signal to extracellular fluid and only has to travel small distance (ex: histamine)
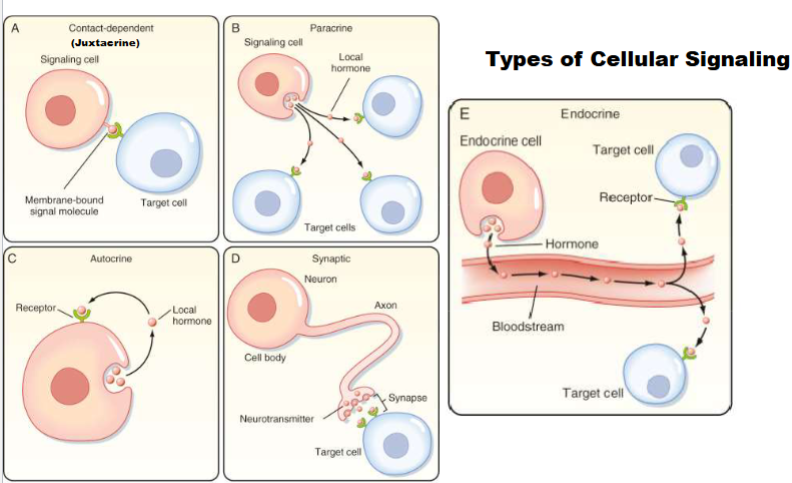
Autocrine (Type of cellular signaling)
single cell both signal cell and target cell, has signal that releases and binds that signal (ex: cancer cells)
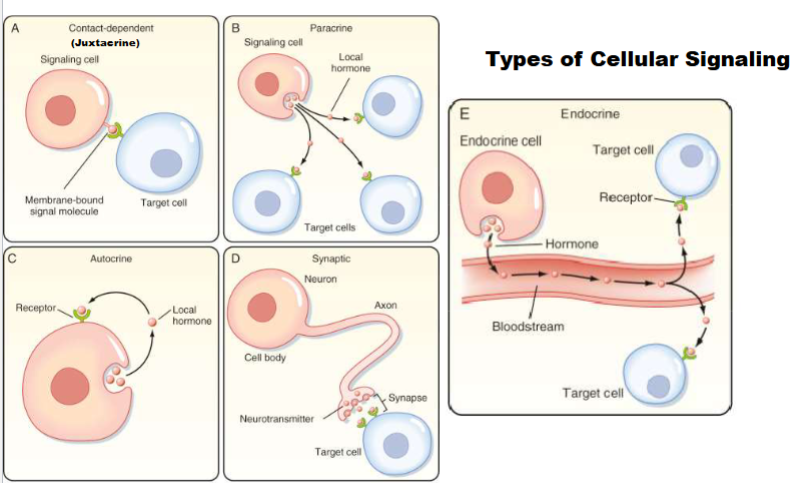
Synaptic (Type of cellular signaling)
signal cell release signal molecule (neurotransmitter), signal has to travel on synapse and target cell has receptor to respond
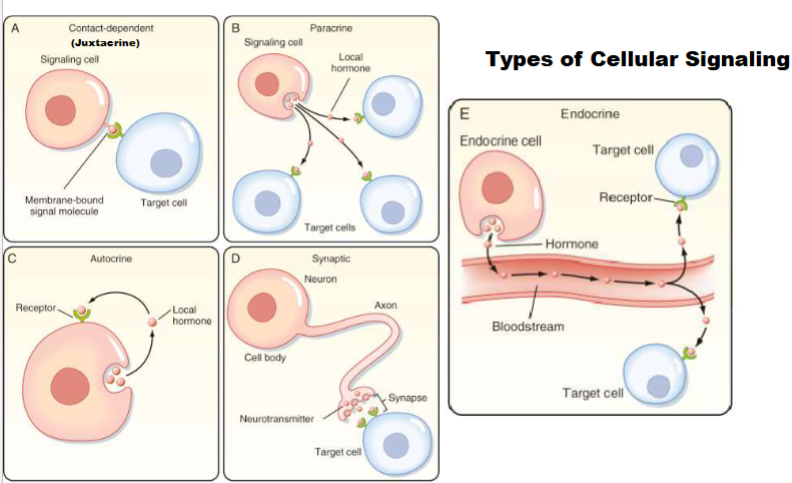
Endocrine (Type of cellular signaling)
signal and target cell are distant from eachother, signal release enter into bloodstream and travel through blood (ex; pituitary gland)
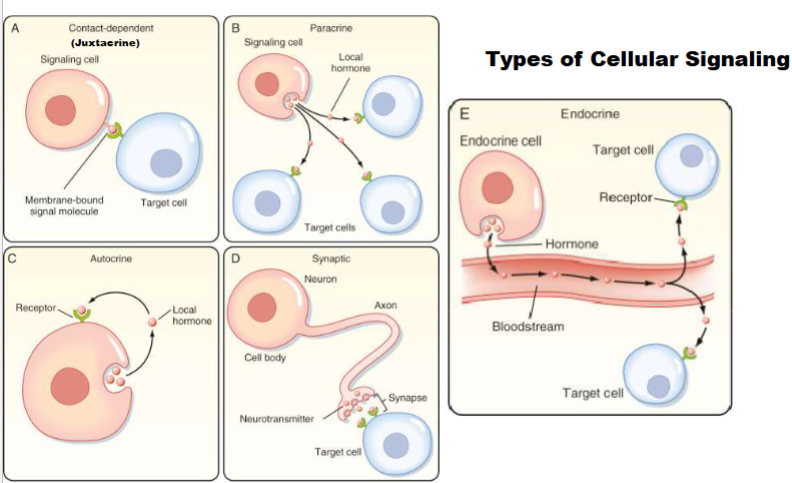
2 Main Receptor Types
Cell Surface Receptor: anchored in plasma membrane of cell
signal transduction: transfe of info.
signalling molecule never makes it inside of cell only goes on surface
Intracellular :signal inside of cell (may in in cytocal )
signal has to find receptor to pass on info.
almost always control gene expression
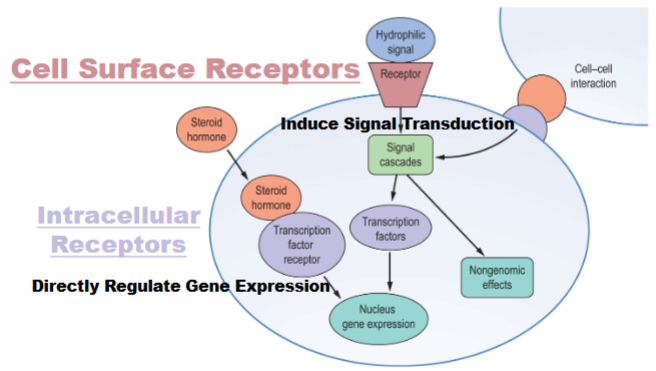
Cell surface receptor signaling
1) start in signalling cell, synthesis molecule, release it and moves through body fluids until it goes to cell
2) signalling molecule bind to receptor, change activity of receptor (change structure and function)
3) receptor receive info. and mod. in some way and mod. internal factors (usually mod other proteins)-signal transduction pathway
4) get target cell gets influenced to do something(change in enzyme activity or gene expression)
5) get rid of signal and shut down signal transduction pathway
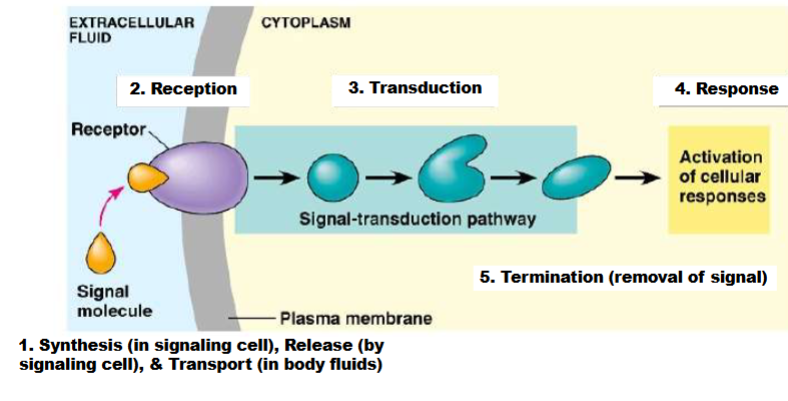
Common Features of Signal Transduction Pathways
2 major types of cellular responses are induced by activation of signal transduction pathways:
A. Regulation of pre-existing proteins
B. Changes in gene expression
1 receptor can activate multiple signal transduction pathways, each of which can elicit different cellular responses
Some Major Types of Cell Surface Receptors pt 1
I. G protein-coupled receptor (GPCR)- most common encountered
A. Single receptor protein associated with heterotrimeric G proteins
B. Utilizes cAMP/PKA & IP3/DAG/Ca2+ pathways
II. Receptor tyrosine kinase (RTK)
A. Multi-subunit complex possessing intrinsic protein tyrosine kinase enzyme activity (i.e., it phosphorylates the Tyr residues of proteins)
B. Utilizes Ras/MAPK, PI3K, JAK/STAT, & IP3/DAG/Ca2+ pathways
Some Major Types of Cell Surface Receptors pt 2
III. Tyrosine kinase-associated receptor (not an enzyme but associated w/)
A. Multi-subunit complex associated with protein tyrosine kinase enzymes
B. Utilizes the same pathways as RTKs
IV. Ligand-gated ion channel
A. Multi-subunit complex that forms a gated pore through the plasma membrane
B. Controls ion flow into & out of cells
Receptor-Ligand Interactions
I. Receptors display binding specificity (1 ligand/receptor; ligands are more promiscuous)
II. The reversible binding of a ligand to receptor is mediated by noncovalent interactions (e.g., H bonding, ionic interactions, hydrophobic interactions, van der Waals forces)
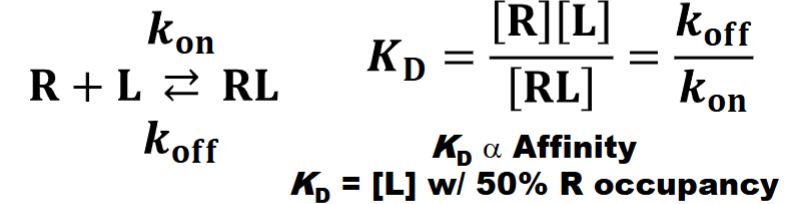
Sensititivity
I. This is the dependence of cellular response on a cell’s receptor # for particular ligand (# receptors determine sensitivity)
A. ↑ receptor #, ↑ sensitivity to particular ligand (& vice versa)
II. Regulation of receptor # & expression are key for controlling physiological & developmental event
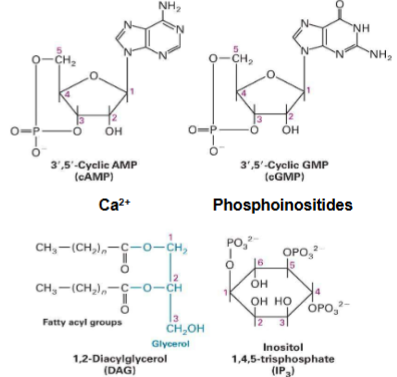
1st Messengers to 2nd Messengers to Signal Transduction Proteins (Intracellular Signal Transduction)
I. 1st messengers = extracellular signaling molecules
II. 2nd messengers = intracellular signaling molecules (know second messengers on pic)
III. 3 main groups of signal transduction proteins are involved in intracellular signaling
GTPase Switch Proteins (G Proteins)- signal transduction proteins (Intracellular Signal Transduction)
I. Heterotrimeric (large) G proteins
(direct receptor interactions)
II. Monomeric (small) G proteins (indirect
receptor interactions) -tertiary structure only
once does signal activity can no longer be a signaler
types of proteins GEF, GTPase
through own enzyme activity or through GAP can control enzyme activity
2 methods of control enzyme function
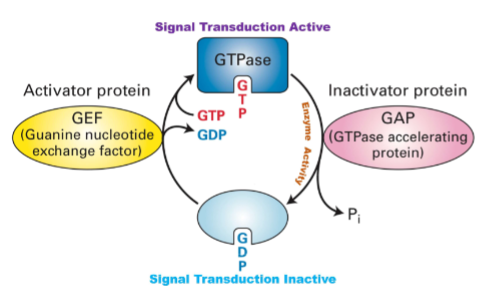
How do GEF, GtPase and GAP effect signal transduction? (Intracellular Signal Transduction)
GEF: Turns the signal switch ON by helping the protein replace GDP with GTP.
GTPase: The protein's own timer that slowly turns the signal OFF by breaking down GTP.
GAP: Speeds up the timer to turn the signal OFF faster.
Summary: GEF turns signaling ON, GTPase slowly turns it OFF, and GAP makes the OFF switch happen quickly to control cell signals precisely
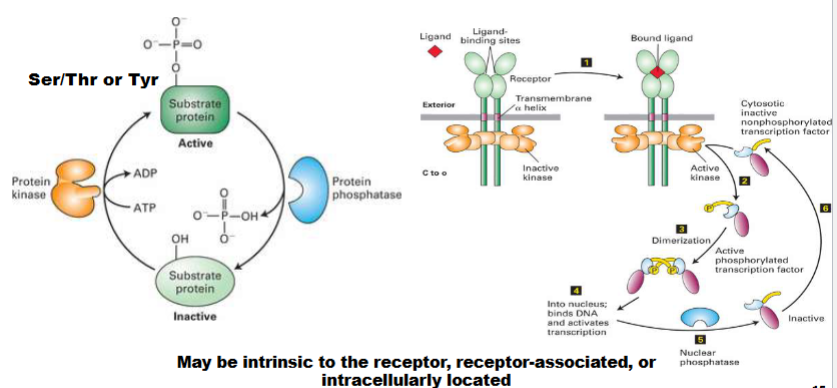
Protein Kinases & Protein Phosphatases (Intracellular Signal Transduction)
kinases add phosphoro groups
Phosphatases remove them
can find this activity in the receptor itself
will tell us is activating or inhibitory
only amino acids that receive these groups: ser/ thr/ tyr
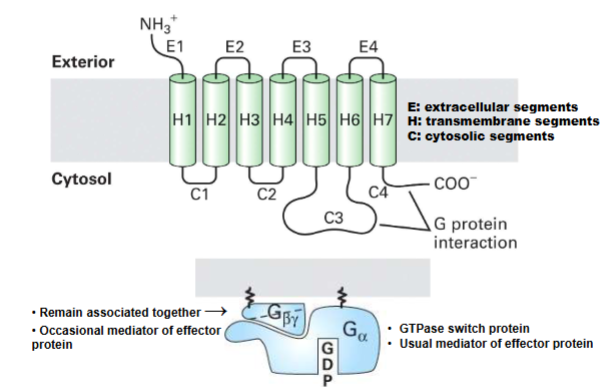
GPCR & Heterotrimeric G protein structure (Basics of G Protein-Coupled Receptors)
top pic: single protein associated w plasma membrane portion of protion on cytocol
extracellar portion where signal
intracellular where g-proteins interact w/
always 7 transmembrane
bottom pic: always remain bound together gbeta gamma
main function is to regulate g alpha
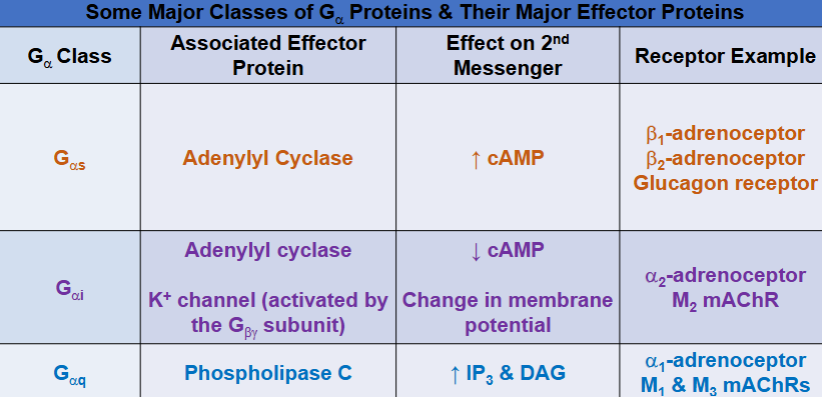
important alpha subunits and effector proteins (Basics of G Protein-Coupled Receptors)
Epinephrine & G alpha s (GPCRs That Regulate Adenylyl Cyclase)
Epinephrine & Galpha(s)
I. Epinephrine (Epi) is hormone
produced by adrenal glands that is
body’s fight-or-flight response
mediator
II. Activates beta1-adrenoceptors (expressed in
heart, kidneys, neurons, & adipocytes) & beta2-
adrenoceptors (expressed in smooth muscle,
liver, heart, skeletal muscle, neurons)
A. Epi → beta1/beta2 → Galphas → AC →
↑[cAMP]Intracellular
Cholera
Disease: Severe, watery diarrhea → can cause death via rapid dehydration.
Cause: Vibrio cholerae
Produces cholera toxin (CT) in the small intestine.
Mechanism:
Toxin enters intestinal mucosal cells → ADP‑ribosylates Gαs protein.
Activation of AC, but
inhibits its GTPase activity
b) Causes continuous cAMP production —> excessive water & electrolyte secretion into
intestinal lumen —> diarrhea
Epinephrine and Gαi (GPCRs That Regulate Adenylyl Cyclase)
I. Epi activates α2-adrenoceptors (expressed in neurons,
pancreas, platelets, ciliary epithelium, & smooth muscle)
A. Epi → α2 → Gαi ⊣ AC (↓ [cAMP]Intracellular)
Whooping Cough
Acute inflammation of upper respiratory tract causing severe coughing attacks, inspiratory whoop, & fainting & vomiting after coughing
A. Caused: bacterium Bordetella pertussis
1. Produces & secretes pertussis toxin (PT)
in upper respiratory system
i. Internalized into upper respiratory cells
where it does ADP-ribosylation of G αi
a) Keeps it in GDP-bound state,
preventing it from inhibiting AC
b) Leads to continuous cAMP production
Protein Kinase A(pka) (GPCRs That Regulate Adenylyl Cyclase)
-each binding makes the next binding easier
when bound proteins will be phorsphoralated
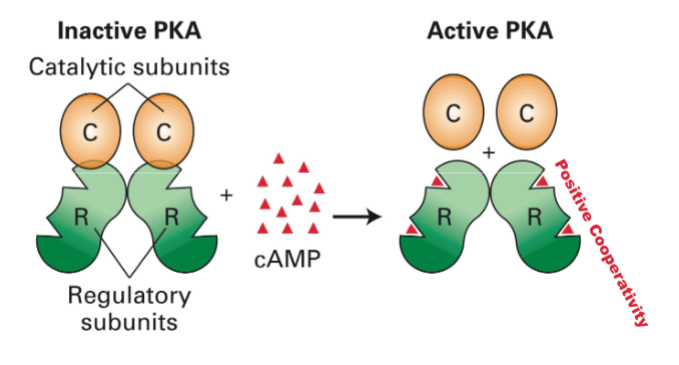
Protein Kinase A
targets for pka is in cytosol, can also go into nucleus
know what pic mean
PKA reg. of GLycogen Metabolism (GPCRs That Regulate Adenylyl Cyclase)
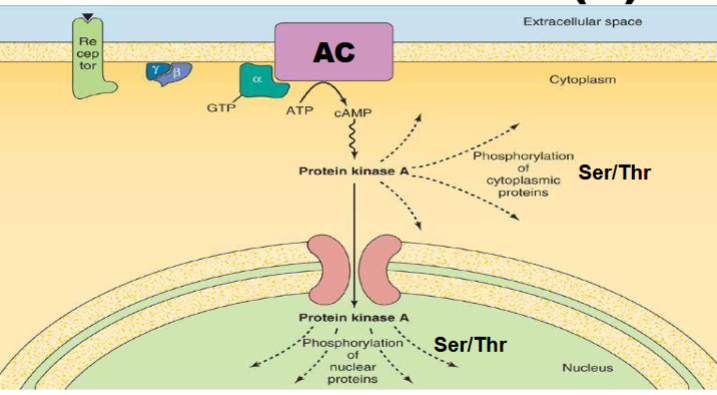
Extracellular signal: Epinephrine or glucagon binds to a G protein-coupled receptor (GPCR) on the cell surface.
Signal transduction: GPCR activates a G protein, which then activates adenylate cyclase (AC) → increases cAMP levels.
cAMP activates PKA: cAMP binds to and activates protein kinase A (PKA).
PKA phosphorylates:
Glycogen phosphorylase kinase (GPK): Activates glycogen breakdown.
Glycogen synthase(GS): Inactivates, blocks glycogen synthesis.
Inhibitor protein(IP): turned on Inhibits PP, maintains enzyme phosphorylation and prevents glycogenesis
![<ul><li><p><strong>Extracellular signal:</strong> Epinephrine or glucagon binds to a <strong>G protein-coupled receptor (GPCR)</strong> on the cell surface.</p></li><li><p><strong>Signal transduction:</strong> GPCR activates a G protein, which then activates <strong>adenylate cyclase (AC)</strong> → increases <strong>cAMP</strong> levels.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>cAMP activates PKA:</strong> cAMP binds to and activates <strong>protein kinase A (PKA)</strong>.</p></li><li><p><strong>PKA phosphorylates:</strong></p><ul><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Glycogen phosphorylase kinase (</span><strong>GPK):</strong> Activates glycogen breakdown.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Glycogen synthase</span><strong>(GS):</strong> Inactivates, blocks glycogen synthesis.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Inhibitor protein</span><strong>(IP)</strong>: turned on Inhibits PP, maintains enzyme phosphorylation and prevents glycogenesis</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/04460699-2d2e-4b90-a864-9f37cc832cfe.png)
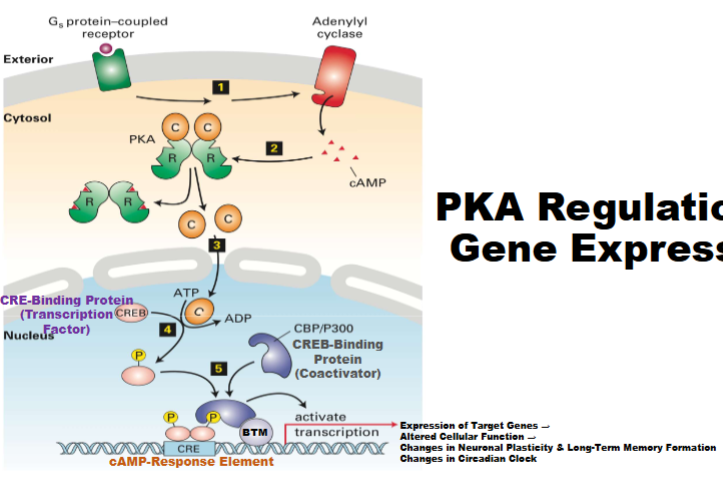
Pka Regulation of Gene Expression (GPCRs That Regulate Adenylyl Cyclase)
catalytic subunits active then move to nucleus target CRE-B (controls gene expression), CRE-B stimulated, will bind to specific sequences of DNA (CRE), when bound can recruit BTM
cytoplasmic as well as nucleic (should know)
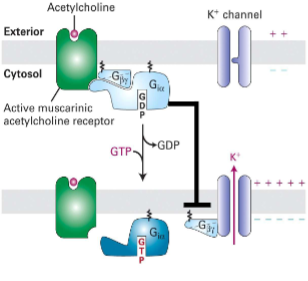
M2 mAChR (GPCRs That Regulate Ion Channels)
I. Muscarinic (responsive to
muscarine) vs. nicotinic (responsive
to nicotine) acetylcholine
receptors (AChRs)II. M2 mAChR (GPCR expressed in
heart, neurons, & smooth muscle) is
activated by AChIII. G-alpha i inhibits AC; activation of
K+ channel by Gbeta y(beta gamma) causes long hyperpolarization of
cardiac muscle cells, slowing
rate of heart contraction(looking at M2 in pic)
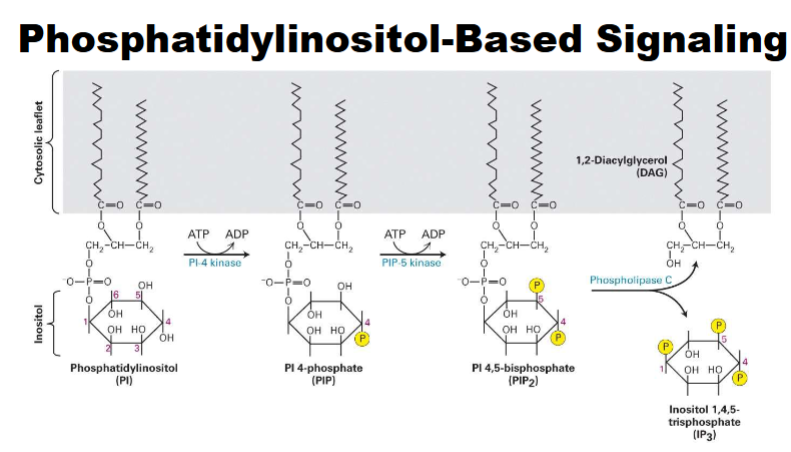
Phosphatidylinositol-Based Signaling (GPCRs That Regulate Phospholipase C)
-phospha. lipids can under go phosphoralyation → PIP→PIP2→ can do to get phospholipase C
DAG imobile (know second messengers) *IP3 mobile
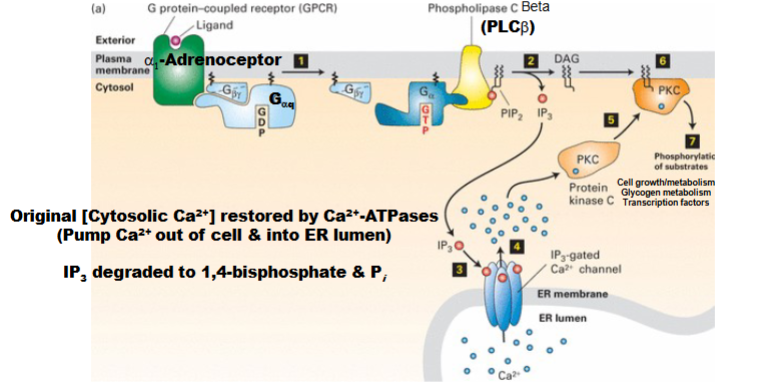
IP3/DAG Pathway & Ca2+/PKC (second messengers) (GPCRs That Regulate Phospholipase C)
Ligand binds GPCR → activates Gαq subunit.
Gαq activates PLCβ → produces IP3 and DAG (second messengers).
IP3 opens IP3-gated Ca2+ channels (normally closed), releasing Ca2+ from ER.
Ca2+(second messenger) and DAG activate PKC:
Ca2+2+ causes PKC to move to membrane and bind DAG → PKC activated.
PKC activation: leads to changes in glycogen metabolism.
Shut-off mechanisms:
Remove Ca2+2+ (via Ca2+2+ ATPase pumps: out of cell or into ER).
Degrade IP3 (returns to 1,4-bisphosphate form)
Ca2+ as a 2nd Messenger (dont memorize) (GPCRs That Regulate Phospholipase C)
just gives idea what Ca does
Ca2+/ Calmodium (GPCRs That Regulate Phospholipase C)
I. Calmodulin is cytosolic protein that binds 4
Ca2+ with positive cooperativityII. Ca2+/calmodulin complex controls activity of
many factorsA. E.g., it activates cAMP/cGMP PDEs
B. E.g., it activates calcineurin
1. Phosphatase that activates T-cell-
specific transcription factor NFAT (important for T-cells for growth and funciton)
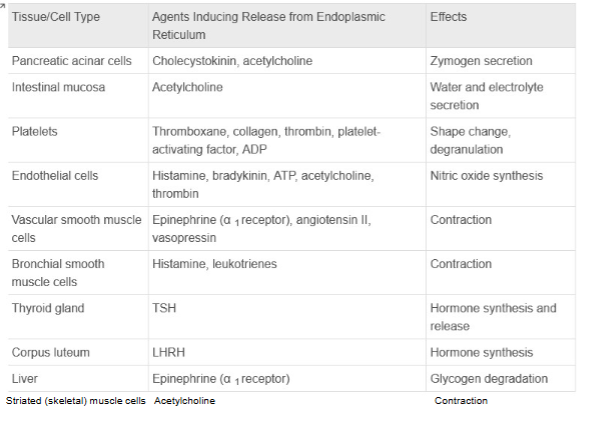
Ca2+/Calmodulin & Myosin Light-Chain (MLC) Kinase (GPCRs That Regulate Phospholipase C)
Stuff on top shows activation of receptor and CA release
Cal. binds to MLC kinase, when unbound its inactive when bound to Ca2+ will become active will form Myosin light chains
Ca2+/Calmodulin & Gene Expression (GPCRs That Regulate Phospholipase C)
when activating or inhibiting will control action of CRE-B
anothe rmecahnism where gene transcription can be controlled
Ca²⁺/Calmodulin Kinase Pathway:
Increased Ca²⁺ activates CaMK II/IV → phosphorylates CREB.cAMP/PKA Pathway:
Increased cAMP activates PKA → phosphorylates CREB (e.g., at serine-133).Transcription Activation:
Phosphorylated CREB binds CRE (cAMP Response Element) on DNA, recruits CBP (coactivator), and initiates gene transcription.Cellular Effects:
Alters gene expression, leading to neuronal plasticity, long-term memory formation, and circadian rhythm regulation.
![<p>when activating or inhibiting will control action of CRE-B</p><p>anothe rmecahnism where gene transcription can be controlled</p><ol><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span><strong>Ca²⁺/Calmodulin Kinase Pathway:</strong></span><br><span>Increased Ca²⁺ activates CaMK II/IV → phosphorylates CREB.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span><strong>cAMP/PKA Pathway:</strong></span><br><span>Increased cAMP activates PKA → phosphorylates CREB (e.g., at serine-133).</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span><strong>Transcription Activation:</strong></span><br><span>Phosphorylated CREB binds CRE (cAMP Response Element) on DNA, recruits CBP (coactivator), and initiates gene transcription.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span><strong>Cellular Effects:</strong></span><br><span>Alters gene expression, leading to neuronal plasticity, long-term memory formation, and circadian rhythm regulation.</span></p></li></ol><p><br></p>](https://knowt-user-attachments.s3.amazonaws.com/bacf269f-9f1c-4bfa-9d66-d60ec9c3ad3f.png)
Ca2+/Calmodulin & Blood Vessel Diameter (GPCRs That Regulate Phospholipase C)
endo. cell will be stimulated by aceytl. receptor will activate Galpha Q and activate second messengers
NO2 activated by__ but acyetl. activates it
NO2 signaling molecule
PDE5 (prevents vasodialation from occuring)
-ACh binds M3 receptor on endothelial cells.
Activates PLC → increases IP₃ → raises intracellular Ca²⁺.
Ca²⁺/calmodulin activates endothelial nitric oxide synthase(eNOS) → produces NO from arginine.
NO diffuses to smooth muscle → activates soluble guanylyl cyclase (sGC) → converts GTP to cGMP.
cGMP activates PKG → lowers Ca²⁺, inhibits actin/myosin → smooth muscle relaxation/vasodilation.
Phosphodiesterase 5(PDE-5) degrades cGMP; inhibition of PDE-5 prolongs vasodilation.
![<ul><li><p>endo. cell will be stimulated by aceytl. receptor will activate Galpha Q and activate second messengers</p></li><li><p>NO2 activated by__ but acyetl. activates it</p></li><li><p>NO2 signaling molecule</p></li><li><p>PDE5 (prevents vasodialation from occuring)</p></li></ul><ol><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>-ACh binds M3 receptor on endothelial cells.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Activates PLC → increases IP₃ → raises intracellular Ca²⁺.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Ca²⁺/calmodulin activates endothelial nitric oxide synthase(eNOS) → produces NO from arginine.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>NO diffuses to smooth muscle → activates soluble guanylyl cyclase (sGC) → converts GTP to cGMP.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>cGMP activates PKG → lowers Ca²⁺, inhibits actin/myosin → smooth muscle relaxation/vasodilation.</span></p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><span>Phosphodiesterase 5(PDE-5) degrades cGMP; inhibition of PDE-5 prolongs vasodilation.</span></p></li></ol><p></p>](https://knowt-user-attachments.s3.amazonaws.com/377451de-1dbf-4815-ba48-69122275a15f.png)
Non-GPCR Cell Surface Recptor Signaling (Non-GPCR Cell Surface Receptors that Control Gene Expression)
Cell signaling primarily controls gene expression.
Multiple ligands, receptors, signaling pathways, and transcription factors are involved.
The same receptor can activate different pathways in different cells, causing varied cellular responses.
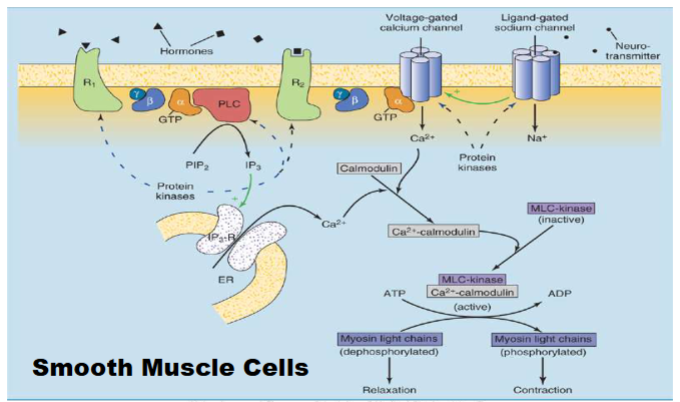
Receptor Tyrosine Kinase (2 types of receptors) (Non-GPCR Cell Surface Receptors that Control Gene Expression)
Step 1: RTKs inactive without ligand (dimerization happens monomers come together)
Step 2: Ligand binding → RTK dimerization.
Dimerized RTKs cross-phosphorylate activation lip tyrosines.
Step 3: Further tyrosine phosphorylation fully activates receptor.
Enzyme’s main targets: themselves (autophosphorylation) → enables downstream signaling
also enzymes
RTK Ligands & Receptors (ligand & receptor share the same name) (Non-GPCR Cell Surface Receptors that Control Gene Expression)
I. Epidermal Growth Factor (EGF) & EGFR
II. Nerve Growth Factor (NGF) & NGFR
III. Platelet-Derived Growth Factor (PDGF) &
PDGFR
IV. Fibroblast Growth Factor (FGF) & FGFR
V. Insulin & Insulin Receptor (IR)
VI. Insulin-Like Growth Factor (IGF) & IGFR
Know highlighted ones
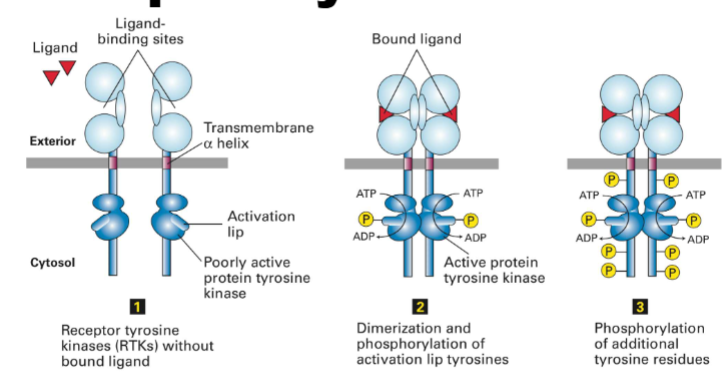
RTK Docking/Adaptor Proteins (Non-GPCR Cell Surface Receptors that Control Gene Expression)
qinternary structure ex.
know IRS-1 (first proteins going to insulin-add rest)
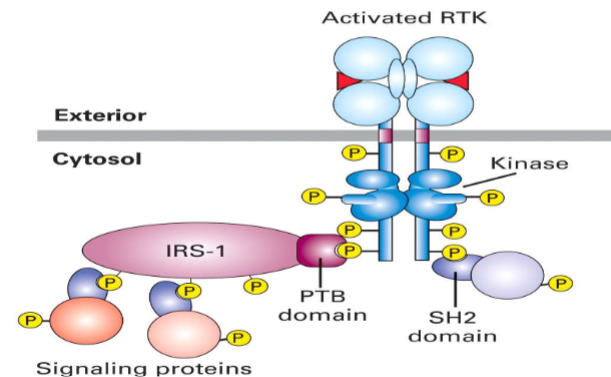
Ras-Mitogen-Activated Protein Kinase (MAPK) Signaling (Non-GPCR Cell Surface Receptors that Control Gene Expression)
Growth factor binds RTK → dimerization and autophosphorylation.
Adapter proteins activate Ras (G protein).
Kinase cascade:
Raf (MAP3K/MAPKKK) → MEK (MAP2K/MAPKK) → MAPK (ex Erk, JNK, p38).
Activated MAPK enters nucleus → activates transcription factors (Fos, Jun, Myc) → changes gene expression.
Main effects: cell growth, division, survival, inflammation, cell cycle progression (mitosis).
Cancer relevance: pathway often overactive due to mutated Ras, driving uncontrolled cell division.
![<ul><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Growth factor binds RTK</strong> → dimerization and autophosphorylation.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Adapter proteins activate Ras</strong> (G protein).</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Kinase cascade:</strong></p><ul><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Raf (MAP3K/MAPKKK)</strong> → <strong>MEK (MAP2K/MAPKK)</strong> → <strong>MAPK</strong> (ex Erk, JNK, p38).</p></li></ul></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Activated MAPK enters nucleus</strong> → activates transcription factors (Fos, Jun, Myc) → changes gene expression.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Main effects:</strong> cell growth, division, survival, inflammation, cell cycle progression (mitosis).</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>Cancer relevance:</strong> pathway often overactive due to mutated Ras, driving uncontrolled cell division.</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/c906a03f-8a79-4da0-92db-6de7633a7068.png)
RTK & PLC
RTK activation leads to phosphorylation of PLCγ.
Activated PLCγ converts PIP₂ in the membrane into DAG and IP₃.
DAG stays in the membrane and activates protein kinase C.
IP₃ diffuses in the cytoplasm and triggers Ca²⁺ release from the ER
![<ul><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2">RTK activation leads to phosphorylation of PLCγ.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2">Activated PLCγ converts PIP₂ in the membrane into DAG and IP₃.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>DAG</strong> stays in the membrane and activates protein kinase C.</p></li><li><p class="my-2 [&+p]:mt-4 [&_strong:has(+br)]:inline-block [&_strong:has(+br)]:pb-2"><strong>IP₃</strong> diffuses in the cytoplasm and triggers Ca²⁺ release from the ER</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/b3c08f44-25e6-47c1-bcd9-0694b16c3a5c.png)
PI3K pathway
targets lipid molecules
-Growth factor binds RTK → receptor autophosphorylation.
-RTK activates PI3K (phosphoinositide 3-kinase)..
-PI3K converts PIP₂ to PIP₃ (second messenger) at plasma membrane.
-PIP₃ recruits proteins for cell growth, survival, and metabolism.
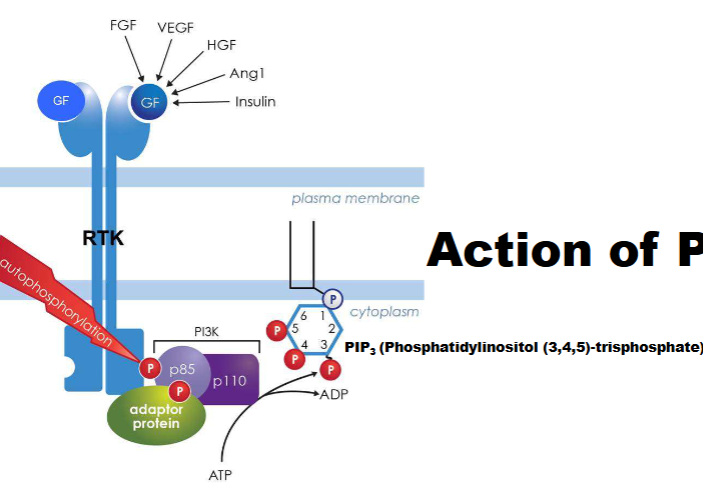
PI3K & AKt/PKB
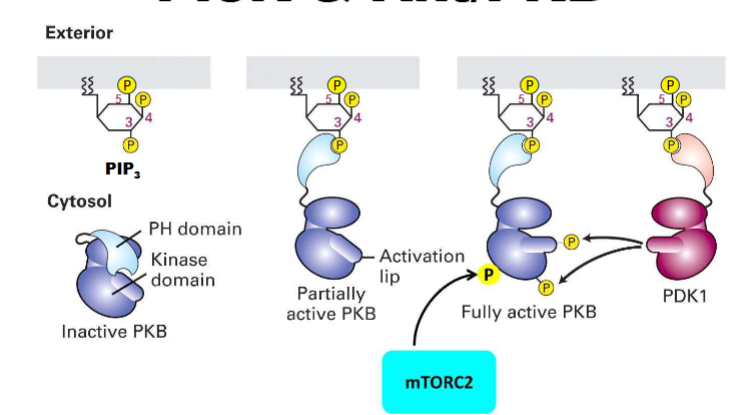
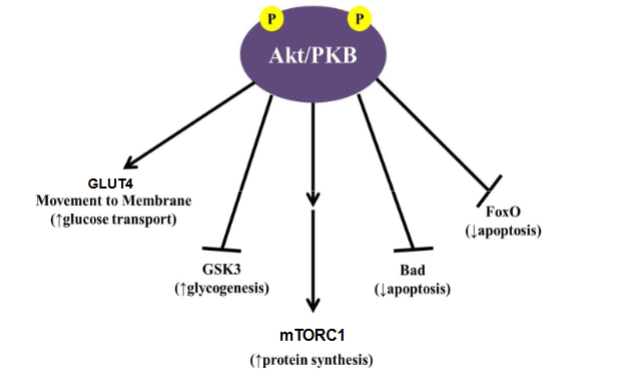
AKt/PKB Signaling
know GLUT4, GSK3, mTORC1,(stimulate protein synthesis), Bad + FoxO (inhibit apoptosis)
it phosporylates each of these
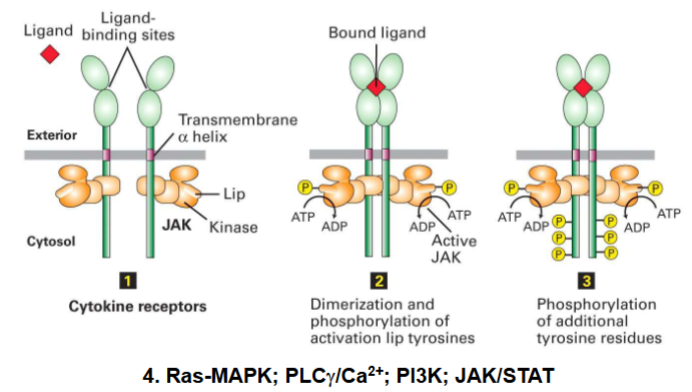
Tyrosine Kinase-Associated Recptor
Cell surface receptor (cytokine receptor type):
Intracellular domain associates with protein tyrosine kinases
Activated by cytokines (immune signaling molecules)
Examples: EPO, G-CSF, IFNs, ILs
Signal transduction via JAK (Janus kinase) enzymes
Cytokine Receptor Signaling
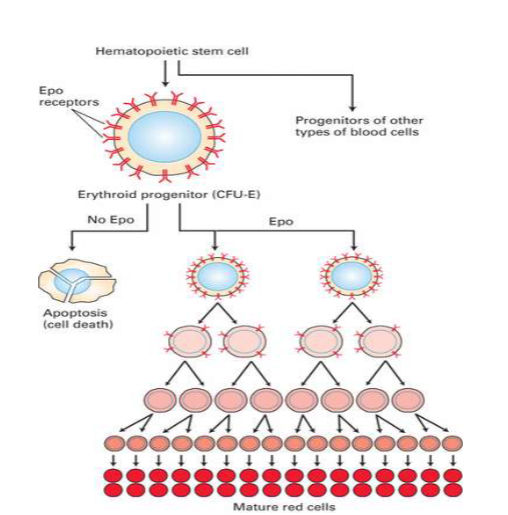
EpoR
I. EPO is produced & secreted by renal cortex in response to ↓[O2]Blood
II. Activates EPO receptor (EpoR) on erythroid progenitor cells
A. Activates JAK2/STAT5 pathway
1. STAT5 (transcription factor) ↑ expression of Bcl- XL
i. Bcl-XL: Anti-apoptotic protein that stimulates proliferation & differentiation of erythroid progenitor cells into mature RBCs 52
![<ul><li><p>I. EPO is produced & secreted by renal cortex in response to ↓[O2]Blood </p></li><li><p>II. Activates EPO receptor (EpoR) on erythroid progenitor cells </p><ul><li><p>A. Activates<mark data-color="yellow" style="background-color: yellow; color: inherit;"> JAK2/STAT5</mark> pathway </p><ul><li><p>1. STAT5 (transcription factor) ↑ expression of <mark data-color="yellow" style="background-color: yellow; color: inherit;">Bcl- XL </mark></p><ul><li><p>i. Bcl-XL: Anti-apoptotic protein that stimulates proliferation & differentiation of erythroid progenitor cells into mature RBCs 52</p></li></ul></li></ul></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/82730b81-5183-471b-b952-23c3bf7f8914.png)
Epo pt2
immature red blood cell, when released will go to pathway to keep cells alive and will eventually become mature
without epo cells willl eventually undergo apoptosis
Epo keeps them alive
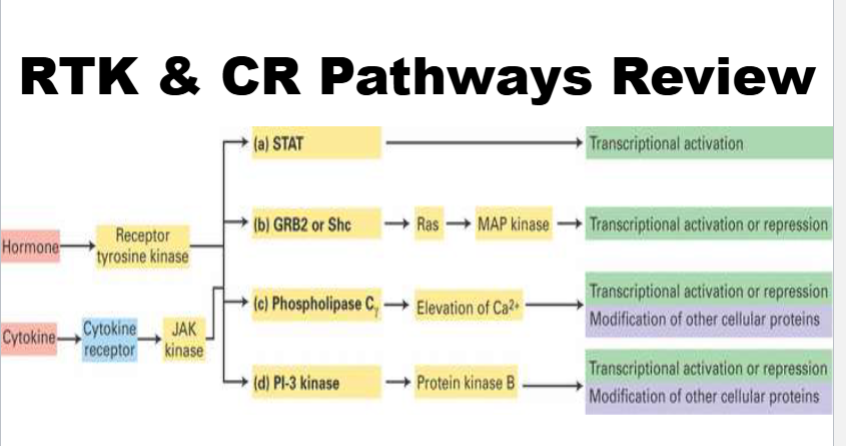
RTK & CR Pathways Review
these pathways are used by rtk (receptor is both receptor and enzyme) and cytokine receptors (receptor is just a receptor)