Amino Acids + Proteins
1/66
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
67 Terms
What is the type of reaction that occurs when amino acids are made into a protein and when a protein is broken down into amino acids
Condensation reaction = amino acids are condensed to form proteins that are linked by peptide bonds
Hydrolysis reaction = breaks up a protein into amino acids
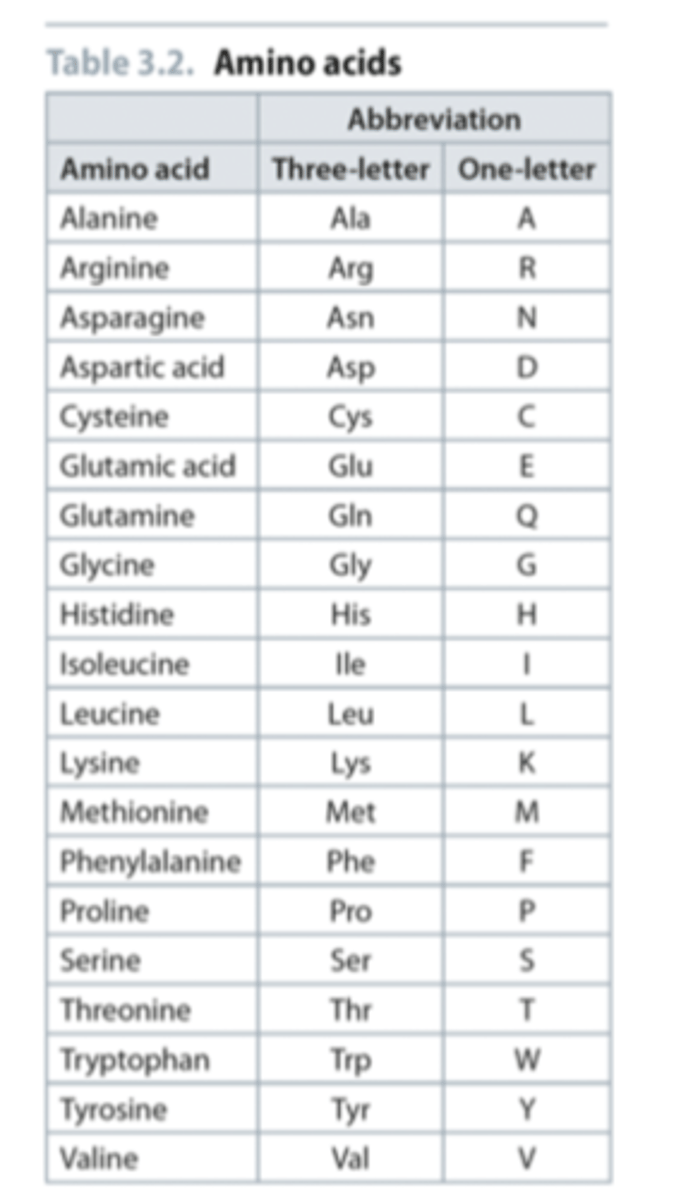
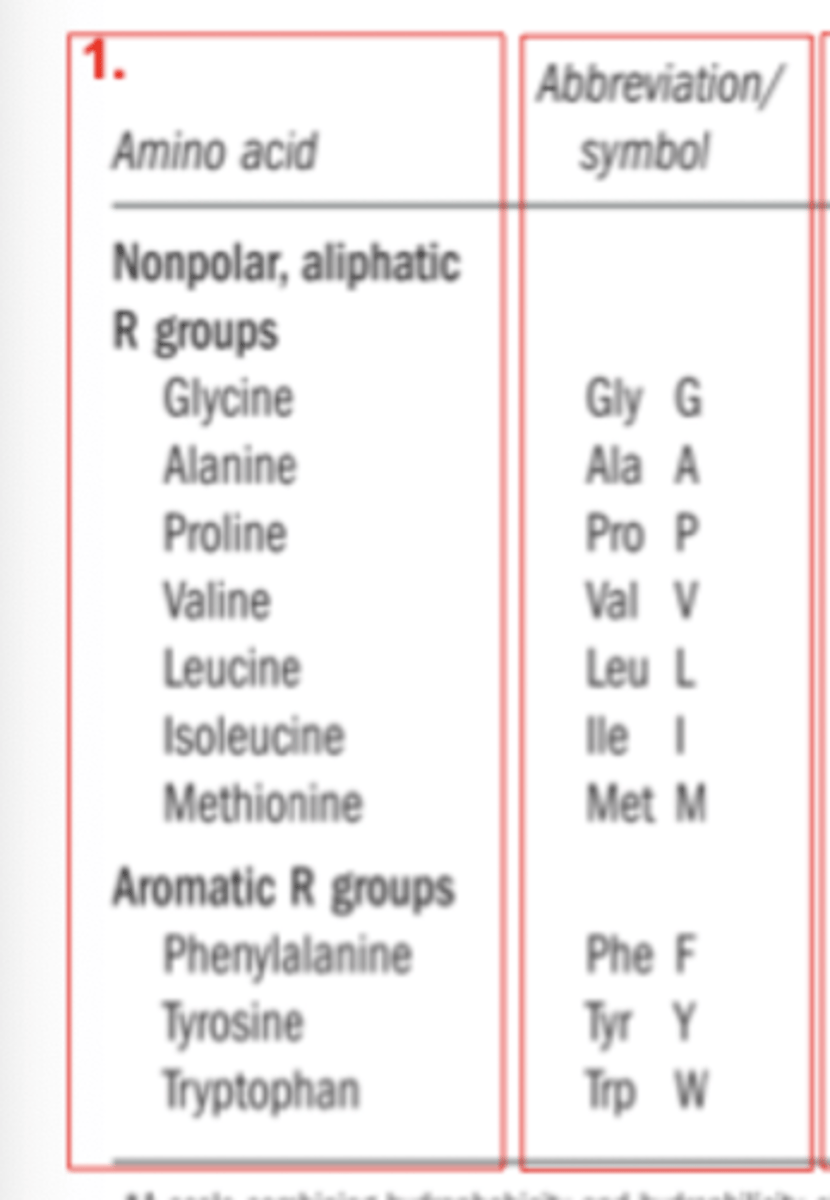
What are the 3 letter and 1 letter abbreviations of the 20 core amino acids
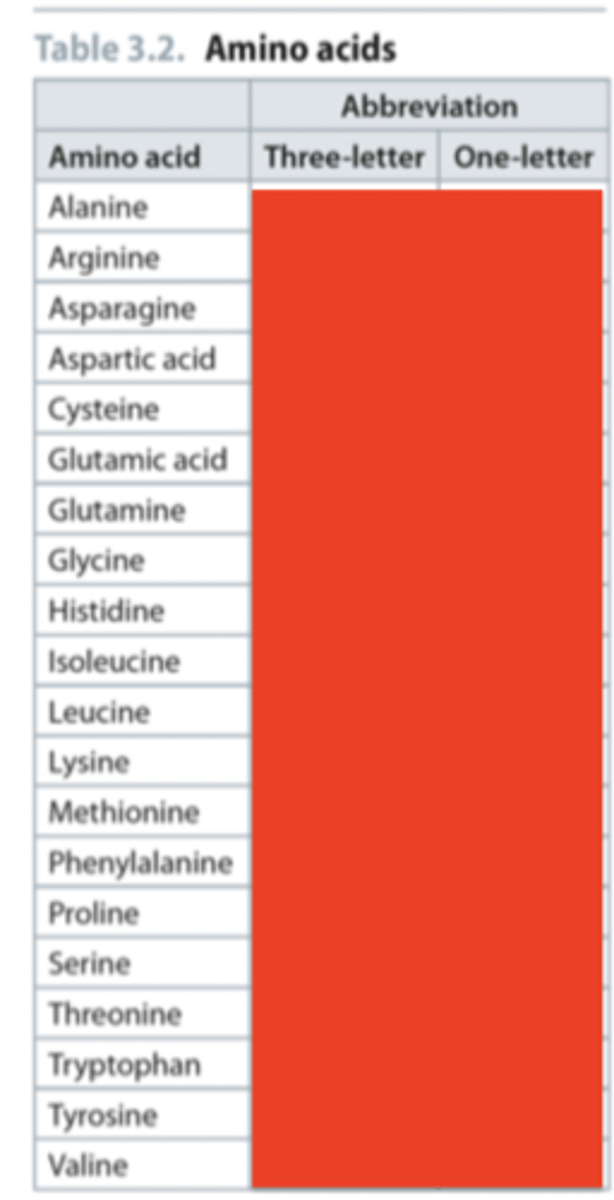
Answer to naming question
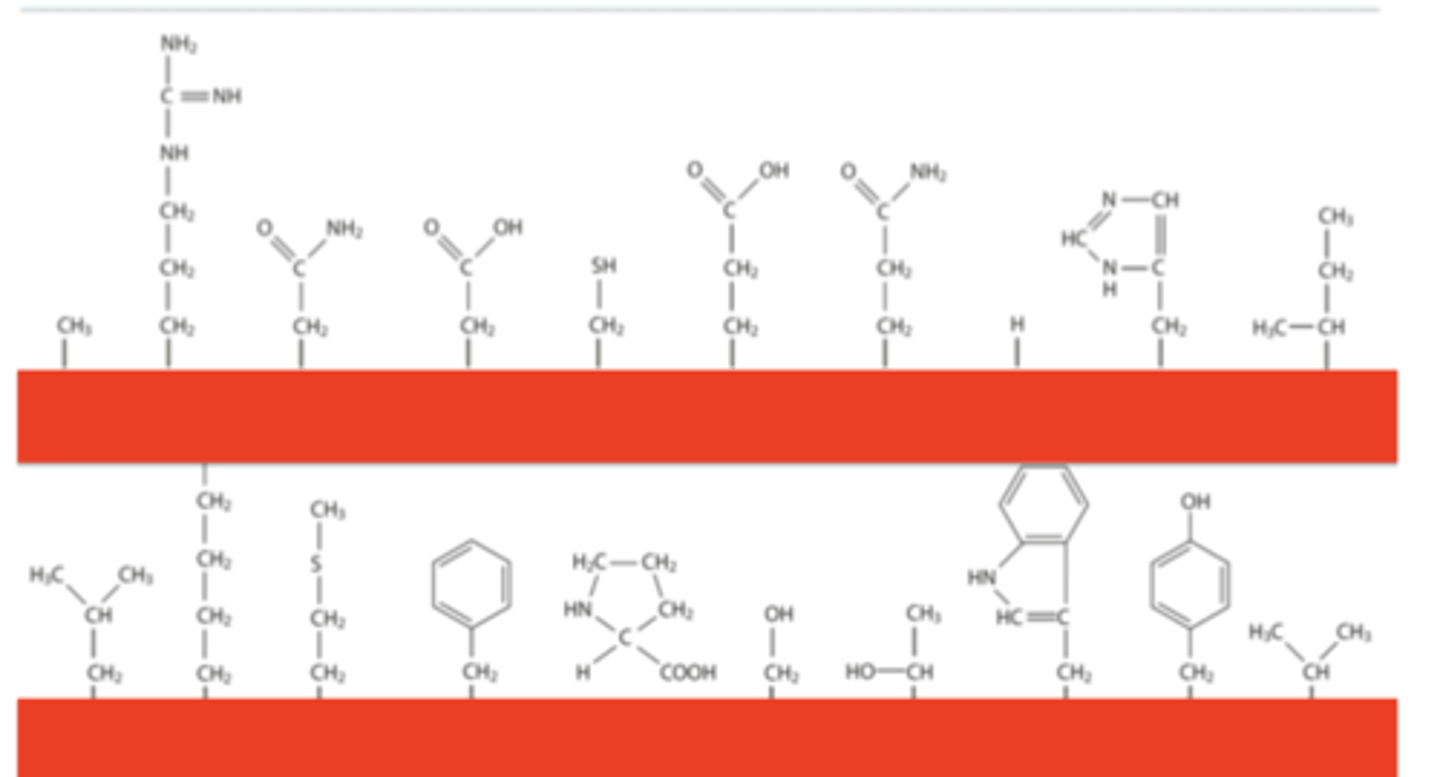
Allocate the amino acid to its R-group
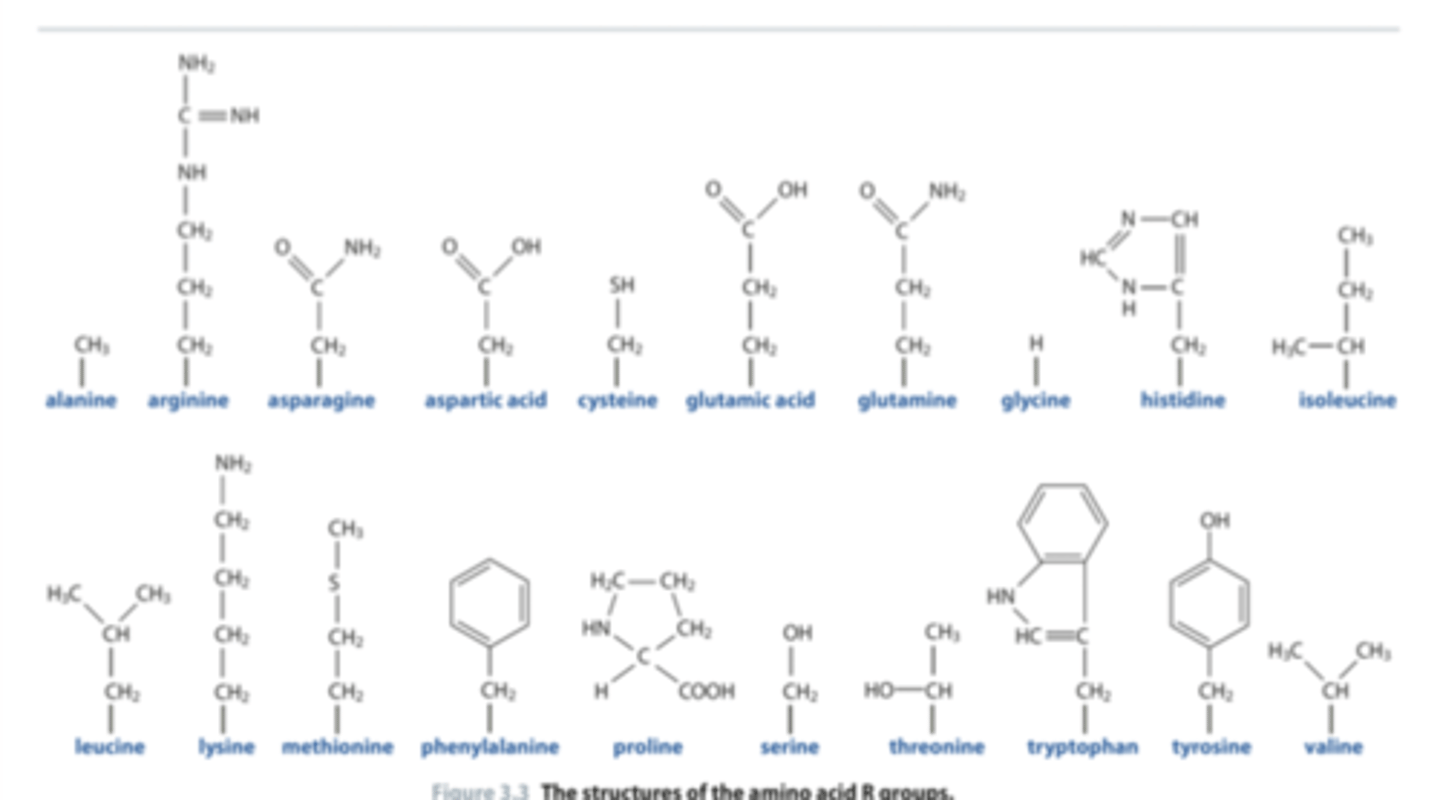
Answer to shapes question
What makes an amino acid R-group polar, non-polar, polar +ve or polar -ve
Polar = has an electronegative atom (can form H-bonds)
Polar +ve = nitrogen atom present that can be protonated
Polar -ve = carbon atom that can lose a proton
Non-polar = no electronegative atoms (cannot form H-bonds)
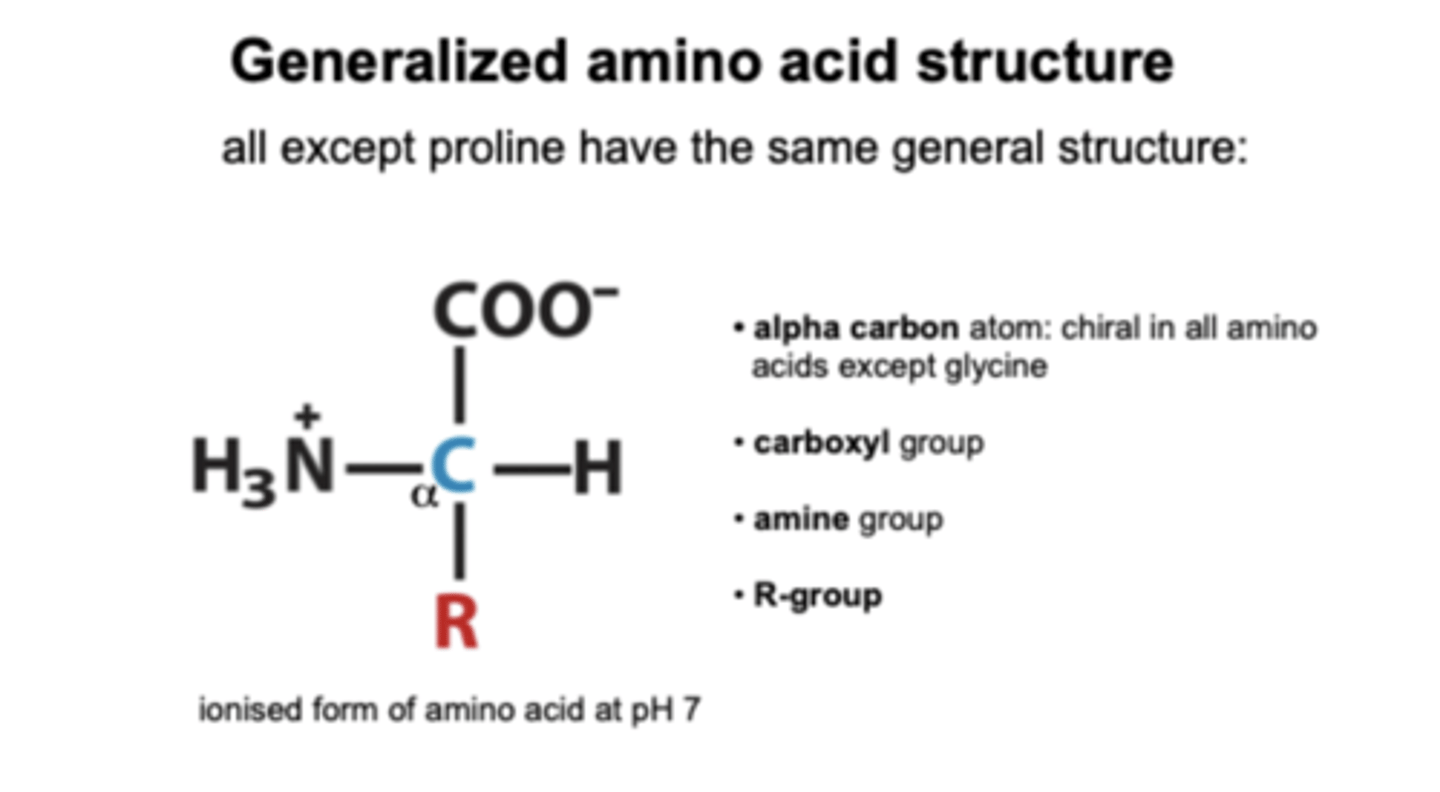
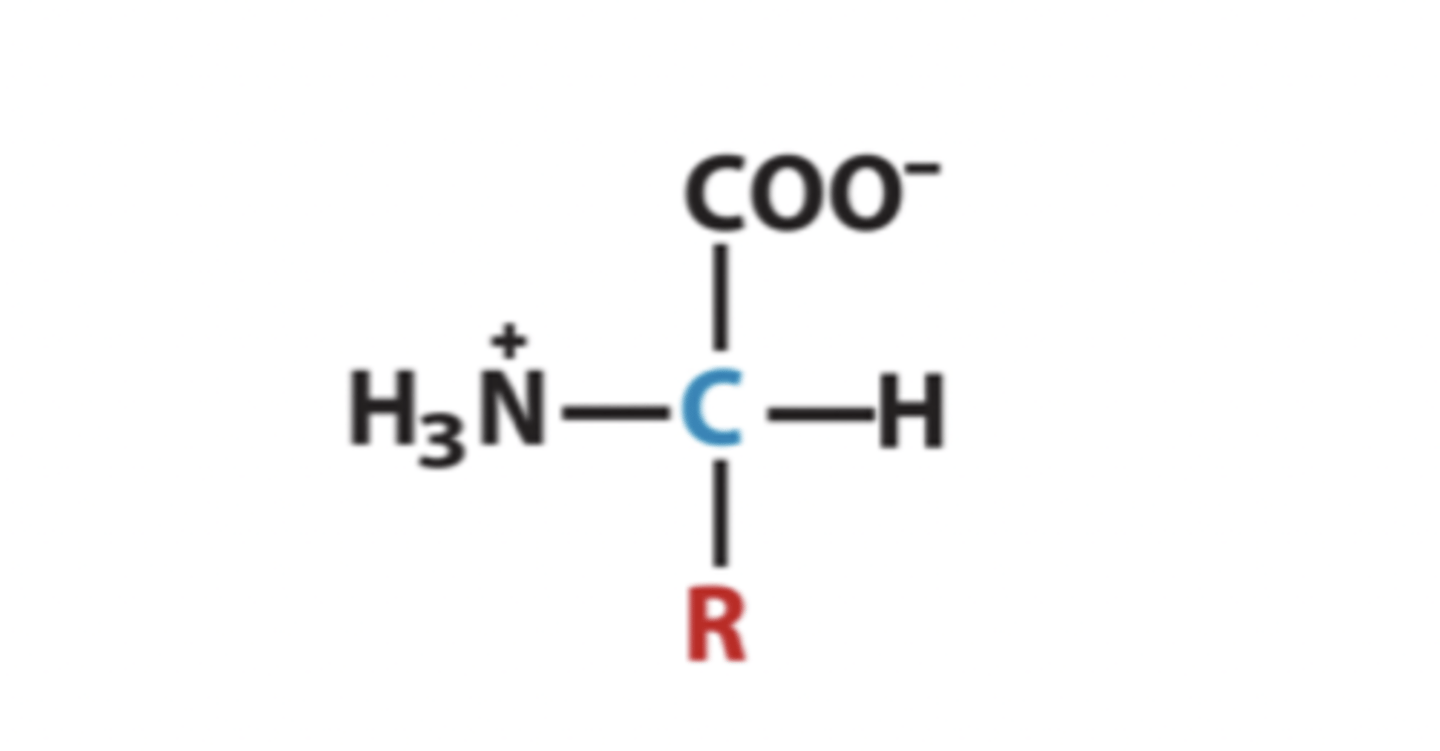
What is the generalised amino acid structure?
R-group gives the protein its functional properties
Name the non-polar aliphatic R group amino acids
- Valine
- Alanine
- Isoleucine
- Leucine
- Methionine
- Proline
- Glycine
Name the aromatic R group amino acids
- Phenylalanine
- Tyrosine
- Tryptophan
Name the polar uncharged R group amino acids
- Serine
- Threonine
- Glutamine
- Asparagine
- Cytosine
Name the polar positive charged R group amino acids
- Lysine
- Histidine
- Arginine
Name the polar negative charged R group amino acids
- Aspartate (aspartic acid)
- Glutamate (glutamic acid)
Explain chirality in terms of amino acids
Amino acids have chirality (each amino acid has 2 forms which are mirror images of each other as the alpha carbon has 4 different functional groups attached) EXCEPT GLYCINE (2 H attached)
This means they form optical isomers (enantiomers)
What are the names of the two optical isomer forms and how are amino acid proteins found
Laevo (L) = left handed
Dextro (D) = right handed
Amino acid proteins are always L-isomers (known as homochirality)
A racemic mixture of D and L enantiomers are formed by chemical synthesis
Which of the following amino acids have nonpolar, aliphatic R groups and which have aromatic R groups
Isoleucine, Tyrosine, Tryptophan, Glycine, Alanine, Methionine, Phenylalanine, Leucine, Valine, Proline
Define the following terms:
- Aliphatic
- Aromatic
- Polar
- Charged
Aliphatic: the R-group chain is NOT branched
Aromatic: the R-group is based on a benzene ring (hydrophobic)
Polar: the R-group carries small charge, both positive and negative (H-bonds)
Charged: the R-group is ionisable creating +ve or -ve charge depending on pH and type of R group (electrostatic/ionic interaction)
Describe the role of pKa in amino acids
Amino acids in solution all have two ionisable groups: carboxyl and amine. These two groups mean that amino acids can both donate and accept protons (amphoteric).
These are not free to act chemically in proteins as they form peptide bonds. HOWEVER, amino acid R-groups may also be ionisable.
pKa is the pH at which a group has equal amounts of protonated and unprotonated forms
Describe the association between proton binding affinity and pKa
Proton binding affinity: how well something takes up protons.
Low pKa = very bad at taking up protons so amino acid will never be positively charged
High pKa = very good at taking up protons so amino acid will be positively charged
What are two amino acids with positively charged carboxyl groups and how does this affect their function
Lysine and Arginine have high pKa so can take in protons easily and have a strong positive charge. Form bonds with negatively charged DNA
Lysine has a strong positive charge at one point
Arginine has branches at positive charge so has delocalised weaker positive charge giving it versatility
What are the 2 amino acids with negatively charged carboxyl groups and how does this affect their function
Aspartic acid and Glutamic acid have low pKa so tend to give protons away so have a negative charge - bind through Hydrogen bond interactions. Aspartate and Glutamate can bind through electrostatic interactions. Bind to magnesium
Ser and The are polar amino acids, what does this allow them to do?
R-groups on these amino acids have O-H bond so H-bonds can form between molecules
What is amino acid pI
The pH at which the amino acid has no net overall charge: zwitterionic. Amino acids are least soluble at the pH that is the pI
What does a high/low hydropathy index imply for an amino acid? What amino acids have high values?
Hydropathy = how hydrophobic (water hating) an amino acid is
High value = highly hydrophobic (Isoleucine is most hydrophobic)
Valine, leucine and methionine are also very hydrophobic
What is the role of the amino acid proline in proteins
Proline is cyclic so kinks the peptide bond - gives the protein structural rigidity
In which amino acid is a thiol group present? What does this signify?
Cysteine can form disulphide bridges with other amino acids in the thiol group. Gives the protein structure by forming these strong covalent bonds
What are the 4 fundamental non-covalent forces of molecular biology
1. Electrostatic interactions (ionic bonds)
2. Polar interactions (H-bonds)
3. Van der Waals interactions (dipole moments)
4. Hydrophobic interactions
What is the importance of the fundamental non-covalent forces
They are crucial for:
1. The chemical reactivity of proteins as enzymes
2. The ability of a protein to fold into the correct shape (conformation) required to fulfil its functions in the cell
What are the benefits of non-covalent forces in comparison with covalent forces?
Non-covalent forces are weak individually but provide proteins with flexibility and ability to move - these bonds can form and break easily and they are in abundance
Covalent forces are stronger but are costly and inflexible
Describe electrostatic interactions between amino acids
These forces occur between atoms that are oppositely charged. They are the strongest forces as they are ionic bonds that form between fully/formally charged atoms. This is outlined in Coulomb's law
Ionic bonds can form and break between 2 R groups which allows for protein movement (conformational change).
Describe an example of an essential ionic bond in enzyme activity
Helicase (a ATPase) hydrolyses an ATP molecule to unwind a double strand of DNA. This reaction occurs due to the ionic bond between lysine and ATP. Presence of lysine holds the ATP in position.
Describe what is meant by polar bonds
A specialised type of polar interaction that involves a slightly electropositive H atom (delta positive) that interact with an electronegative acceptor atom (delta negative). Most H-bonds in proteins occur between H and O/N.
Describe the use of water having hydrogen bonding in protein function
1. Provides a solvation shell around the protein. Water forms a shell around proteins by a network of H-bonding to amino acids. This ensures the protein remains hydrated.
2. Controls the ionic interactions between opposite charges by reducing them
Describe what is meant by Van de Waals forces and their role in amino acids
Random movement of electrons in the electron cloud around an atom causes attraction to the electron clouds of other atoms via small positive and negative charges. However, there is an optimal distance between atoms which allows them to be attracted to each other (too close and they will repel). These interactions are very weak but are found wherever there are atoms.
These forces are what draws two amino acid R groups together which facilitates electrostatic interaction between the groups. So all amino acids have Van de Waals forces.
Describe what is meant by hydrophobic forces
These are forms of energy (not bonds) and are crucial in protein structure and function.
Hydrophobic molecules crowd together in water, by pushing out of the way water molecules. These molecules avoid water as it is not energetically favourable. Oils are non-polar and do not hydrogen bond to water.
Hydrophobic interactions between non-polar molecules are energetically more favourable than the energy required for non-polar molecules to dissolve.
Where do hydrophobic forces occur in amino acids and what are their role
Hydrophobic forces occur in the middle of proteins (hydrophobic core) - the part of the protein that is the furthest away from water. These amino acids pack together repelled from aqueous shell which surrounds them. The energy between the hydrophobic molecules causes the core to be very stable.
Hydrophobic forces increase with temperature and salt.
Define the term polyamide
The functional unit of a protein
Describe the reaction between amino acids to form a peptide
A condensation reaction is the reaction between amino acids that create a peptide bond. The peptide bond forms between a carboxyl group in one amino acid and the amine in another amino acid. These bonds are 3D and R groups are different shapes and sizes so the protein evolves to ensure the R groups are where they are supposed to be.
Explain the charges of peptide bonds and how it affects its functions (resonance hybridization)
The peptide bond is an amide bond (carbon-oxygen and hydrogen-nitrogen bonds) so has the ability to be polar - have delta positive and delta negative charges so the peptide bond (as well as the R groups of the amino acids) can form H-bonds.
The peptide bonds can contribute (through its delta positive and delta negative ) to the ability of the protein to fold. The position of the peptide bond in relation to the R-groups of amino acids means there is a possibility of interactions between the R groups and peptide bonds (important for 2o structures).
Therefore, means the peptide backbone contributes to protein structure and sometimes directly plays a part in catalysis.
Explain the flexibility of the peptide bond
The peptide bond is flexible because amino acids can rotate about the bonds: alpha C-C' (called psi) and N-alpha C (called phi). The peptide bond being flexible gives the protein flexibility. Rotation for amino acid depends on if there is a steric clash between the amino acid R group and the main backbone.
Psi and phi rotation, and steric clash influencing them give 'backbone shape' to the polypeptide.
Proline kinks the peptide bond as its R-group is cyclic (covalently bonded back to itself - covalent imino acid). This is used to engineer proteins.
Explain why protein folding occurs
Chains of amino acids in peptide bonds fold up into proteins by non-covalent forces.
Proteins fold due to the thermodynamics of hydrophobic amino acid interactions which forces those amino acids to the core of the protein (away from water) which gives the structural stability of the core around which all the rest of the proteins fold around (form a solvation shell).
Describe the role of the non-covalent forces in protein folding
Proteins fold into a conformation of lowest energy
- the hydrophobic amino acids will find one another (more energtically favourable). When they find one another they form a core.
- the hydrophilic amino acids then interact with each other through electrostatic attractions, hydrogen bonds and van der Waals interactions.
Describe the use of urea in altering protein folding and the role of protein chaperones
The primary structure of a protein has all the information required for the folding of a protein.
If you want to unfold a protein, expose it to a high concentration of urea. If you then remove the urea, the proteins reforms into its original shape (intrinsically folds).
Protein chaperones have the ability to find proteins that are not quite folded and then help them fold.
Outline the protein structure hierarchy
1. Primary structure: amino acid sequence and motifs
2. Secondary + super-secondary structure: 3D elements formed from folding of discrete peptides
3. Tertiary structure: The overall protein fold and shape
4. Quaternary structure: When proteins bind together non-covalently via all the 4 non-covalent forces
Outline the secondary structure of amino acids and how amino acid composition affects its elements.
Made up three elements:
- alpha helix: 31%
- beta chain/sheet: 28%
- turns: 30%
Some amino acids are predominant in alpha helix (glutamic acid), others are predominant in beta sheets (tryptophan) and others in turns (glycine). This is a result of non-covalent interactions.
Describe how the alpha helix structure arises
- Most commonly occurring protein secondary structure
- Energetically very favourable for any amino acid sequence because it involves peptide backbone interactions, not R-groups
- The H-bonds are key to structure: the nitrogen on the amine group of one amino acid forms a H-bond with the H on the carbonyl of another amino acid which forms the alpha helix structure.
- Within the structure of the alpha helix there are H-bonds between peptide bonds which in turn forms the structure of the helix, but the alpha helix itself has a positive charge at one end and negative charge at another end.
What does the alpha helix amino acid composition depend on?
The composition depends on the position of the helix in the protein.
Hydrophobic residues will be on the inside of the alpha helix and the hydrophilic residues will wrap around.
Therefore, alpha helixes are made by the H-bonds between peptide bonds of nearby amino acids. The R-groups of these amino acids then give the protein a bigger picture character as they are either pointing inside (hydrophobic) or outside (hydrophilic)
Describe the formation of beta sheets
B-strands rely on association with other B-strands for stability. Multiple B-strands form B-sheets
Intermolecular H-bonds form between peptide bonds (N-H and C-O) in amino acids. When multiple polypeptide chains, called beta-strands, align next to each other and are held together by hydrogen bonds between their backbones. These hydrogen bonds form between the N-H group of one amino acid and the C=O group of an amino acid in an adjacent strand, creating a strong, sheet-like structure that can be either parallel or antiparallel.
NOTE: in alpha helix, the H-bonds are cis molecular (intra) and in beta strands the H-bonds are trans molecular (inter) - IN BOTH R GROUPS NOT INVOLVED
Describe protein turns and loops
Protein 3o structures have lots of turns and loops for conformational flexibility and movement. These are the bits that join helices and strands together. They require little steric clash so very small amino acids are required (glycine and alanine). The amino acid R groups undergo non-covalent interactions to form these turns and loops
Describe the helix-turn-helix motif
Very common for DNA binding proteins.
The motif of an alpha helix-turn (e.g. glycine)-alpha helix forms a pincer across the major groove of DNA - allows for the easy binding of proteins to the major groove of DNA. The amino acid R-groups form the H-bonds with DNA - the nitrogenous bases can form H-bonds with these R groups. Therefore, the presence of these H-bonds allow for the reading of DNA
How are alpha helixes, beta sheets and turns drawn?
Alpha = cylinder
Beta = arrows
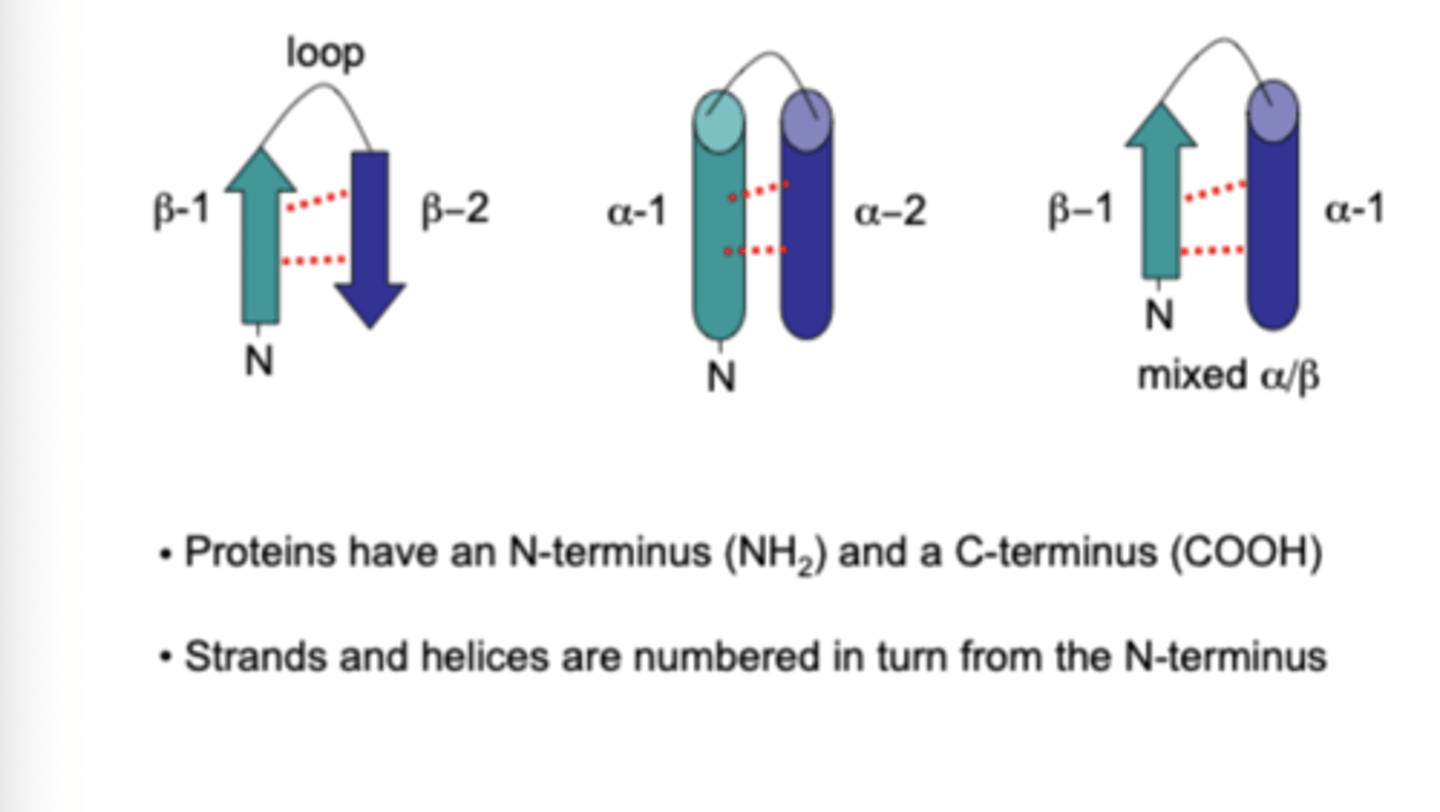
Describe how a super-secondary structure is formed
When a secondary structure comes together to form a super-secondary structure. Most common is the coiled coil - two alpha helices coil around each other as a result of their hydrophobic cores (they pack together to move away from the solvation shells). This packing makes these structures very stable.
Outline the leucine zipper
Leucine zipper is a typical alpha helical coiled coil. The two alpha helices that come together are usually lined with strips of leucine as it is very hydrophobic. The leucine zipper often occurs in DNA binding proteins
Outline the two stranded beta sheet
A small beta sheet lies across the DNA major groove. The beta strands are H-bonded to one another by peptide backbone interactions. This leaves the amino acid R groups free to interact with the DNA backbone and bases.
Describe the arrangement of protein tertiary structures
The 3o structure is the the three dimensional shape of proteins in space. The fundamental unit of a protein's overall 3o structure is a domain. A protein may comprise of one or more domains, each with different roles.
Define the term domain
An autonomously folding peptide chain which provides specific functionality to the structure or function of a protein.
Outline the importance of RecA domains
Any enzyme that needs energy will have a RecA domain.
RecA catalyses homologous recombination which ensures successful DNA replication.
It is an ATPase as it brings two single strands of DNA together by hydrolysing ATP (provides energy).
RecA protein ensures DNA replication and RecA folds are found everywhere for other DNA processes.
Define what is meant by protein motifs
Protein domains contain motifs. Motifs are a conserved amino acid sequence that have the same specific function. The motif sequence can be picked out in proteins from diverse organisms.
Outline amino acid sequence motifs and the most common example
These are a short sequence of amino acids that is recognisable in the primary sequence of many proteins with the same function.
In helicase enzymes (e.g. UvrD) the RecA domain binds ATP via a motif that contains lysine residue. The motif is called a Walker A box and is found in most if not all helicases.
Describe the role of RecA domains + Walker A motifs in UrvD helicase
RecA domain binds to and hydrolyses ATP due to the presence of the Walker A motif. The Walker A motif contains lysine residue that forms a salt bridge to gamma phosphate of ATP holding it in position so it can be cut off to generate energy. Glycine residue is also present because it is very small allowing for very tight folding without amino acids getting in the way. The hydrolysis of ATP allows for the action of helicase on DNA and therefore for DNA replication to occur.
What are structural motifs and the Greek Key
They are energetically favourable ways proteins fold so that the proteins can preform their function. The Greek Keys is an amalgamation of beta strands. They allow proteins to fold in a particular way so that certain bits of proteins can get close to DNA.
Describe the formation of quaternary structures
When proteins bind together non-covalently via all the 4 non-covalent forces. Proteins can interact with other proteins in a number of forms:
- Lots of identical proteins bind together and then can form homomultimers and homodimers
- Lots of different proteins bind together and can form hetermultimers and heterodimers
Describe an example of a homodimer
2 monomers of identical HIV-1 integrase protein enzymes bind to each other forming the HIV-1 virus.
Describe an example of an heterodimer
DNA replication in our bodies is driven by a complex heterodimer known as MCM. MCM is a DNA helicase that forms a ring of 6 different monomers and spins round moving all the way through the DNA thrashing is apart.
Explain how proteins can read DNA sequences
Most DNA proteins target the major grooves of DNA. 3 H-bonds between G-C and 2 H-bonds between A-T can be read by amino acid R groups which are polar (form H-bonds) and are spaced to match the nitrogenous bases such as glutamine and asparagine. DNA helicase then binds to DNA and hydrolyses ATP. This energy derived from ATP hydrolysis causes movement that unwinds the DNA strands.
Proteins form electrostatic bonds to the negatively charged phosphates that make up the back bone of the DNA.
What are the amino acids involved in the movement that unwinds the DNA strands?
In UrvD there are 4 HTHs that make contact with DNA through hydrogen bonding.
There are H-bonds between arginine peptide bonds, 2 glycines stopping steric clash and exposes peptide bond that is delta negative.
Tyrosine residue to come in and wedge its R-group between the bases of DNA unwinding it due to its benzene ring and OH group (H-bonds). Tryptophan has similar structure to purine in DNA so can stack.
How can DNA sequence be recognised by proteins?
Nitrogenous bases of DNA have a H-bond code. So DNA sequences can be recognised due to the specific H-bonds formed by the nucleotide sequence. The physical spacing of the H-bonds between nucleotide bases is recognised by the physical spacing mimicry of amino acid R groups that bind to that structure.
Outline DNA-metal binding by proteins
Walker A motif requires a Walker B motif to do the chemistry.
Walker B known as a DEAD box as the amino acid sequence that makes up the motif always has aspartate (D) and glutamate (E) residues and 2 other amino acid residues.
The amino acid R groups of D and E are negatively charged so can bind to magnesium ions and so allows for the Walker A to hydrolyse ATP.