Taxonimic evidence 3; Phylogenetic inference/ tree building
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
Tree terminology
dichotomy
a branch point where two lineages diverge from a common ancestor
polytomy
more than two lineages descend from a single ancestral lineage
pleisiomorphy
an ancestral character state or a trait that is present in the ancestor of a group and shared by some or all of its descendants
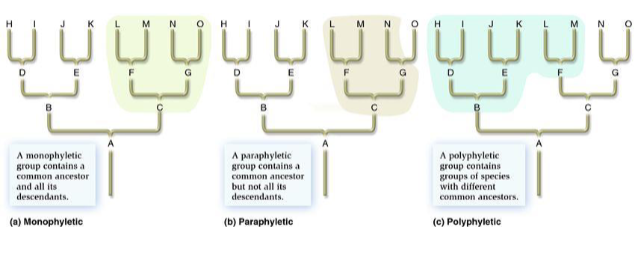
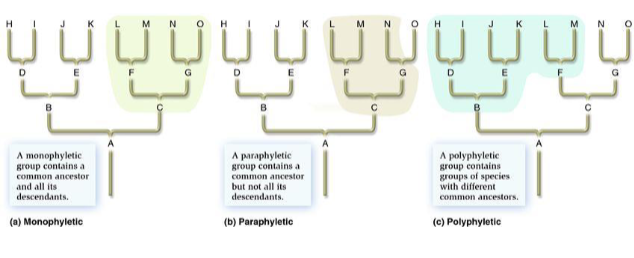
what is a monophyletic group?
a group includ8ing a common ancestor and all of its descendants
a clade
one “cut” from a phylogenetic tree
A group composed of an ancestor and all of its
descendants; diagnosed by synapomorphies, a
clade.
apomorphic
derived characters (not pleisiomorphic)
sympleisiomorphies
shared ancestral characters
not useful
root
common ancestor of whole group
synapomorphy
shared derived characters
topology
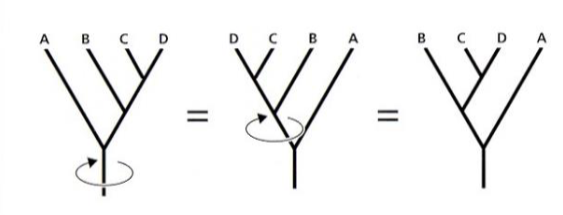
the shape of the tree (the connections between branches)
branches can be rotated at a node, w/out changing relationships among the taxa
paraphyletic group
a group containing a common ancestor and some, but not all, of its descendants; diagnosed by sympleisiomorphies
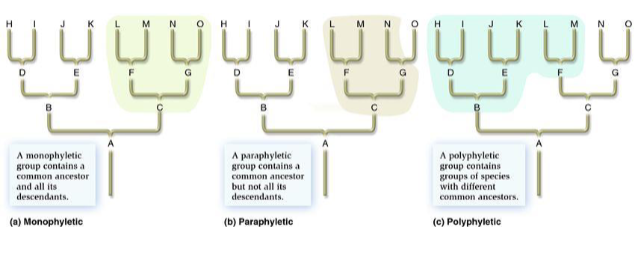
polyphyletic group
a group w/ 2 or more ancestors, but not including the true common ancestors of its members
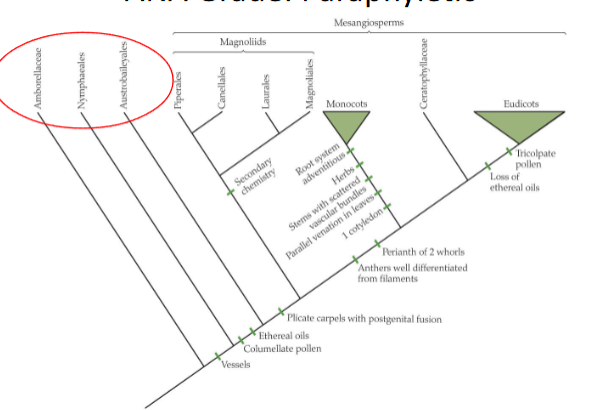
ANA grade is an example of what group
paraphyletic
Phylogenetic inference
1. Obtaining information to develop phylogeny
-kinds of characters
2. Developing a phylogeny
-distinguishing synapomorphies
-rooting the tree with an outgroup
-optimality criteria for constructing trees
-parsimony vs. distance methods
3. Evaluating the phylogeny
-statistical support
-weaknesses of reconstruction methods
4. What do you do with a phylogeny?
developing a phylogeny
distinguishing synapomorphies
rooting the tree with an outgroup
optimality criteria for constructing trees
parsimony v. distance methods
phylogeny reconstruction
The most closely related taxa should share the
most traits (be most similar)
• Relies on the assumption that similarity is due
to shared common ancestry (homology)
• Specifically, relies on synapomorphies: shared
derived traits that characterize a
monophyletic group
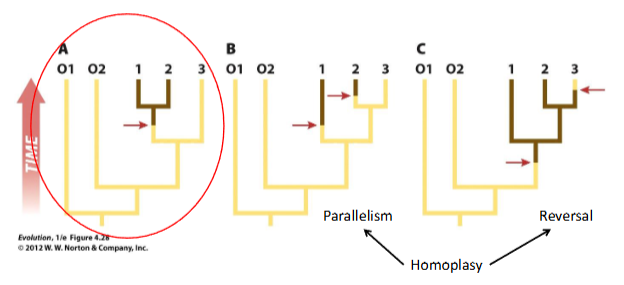
must be distinguished from homoplasy: similarity that is not due to common ancestry
homoplasious traits are parallelisms or reversals: not homologous
what are 2 traits not useful in phylogeny reconstruction
sympleisiomorphy: shared ancestral traits, that are not derived in the group you’re looking at
autapomorphy: a trait that is derived w/in one lineage: not shared with anything else
How do we pick out synapomorphies?
use an outgroup to root the tree
helps to figure out polarity: order of appearance of traits in evolutionary time
assume traits shared between outgroup and ingroup are ancestral
criteria for an outgroup
must assume that all ingroup members are more closely related to one another than the outgroup
outgroup must be separated from ingroup lineage BEFORE the ingroup diversified
3 possible trees for observed character states
Methods of building trees
parsimony (maximum parsimony)
distance methods - mainly neighbor joining
require an explicit model of evolution
Maximum likelihood
Bayesian inference
parsimony is easiest to understand and is intuitively appealing
Phylogenetic inference- parsimony
minimizing the number of evolutionary changes
Occam’s razor: principle that states “the simplest explanation is most likely correct”
parsimony asserts that the tree w/ the least # of evolutionary changes required is the “correct” tree
the only informative characters in parsimony are synapomorphies
no explicit model for evolution
What is the primary difference between a gene tree and a species tree?
A gene tree represents the phylogeny of a single gene, while a species tree represents the phylogeny of an entire species.
What is incomplete lineage sorting?
When gene trees do not reflect species trees due to shared ancestral variation not sorted properly during speciation.
Which of the following methods of tree construction minimizes the number of evolutionary changes?
Parsimony
In a phylogenetic tree, what does a monophyletic group consist of?
A group composed of a common ancestor and all of its descendants.
Explain the difference between a synapomorphy and a homoplasy.
A synapomorphy is a shared derived trait that is used to define a monophyletic group. It indicates a common ancestry. A homoplasy, on the other hand, is a trait shared by species due to parallel evolution or evolutionary reversals, not common ancestry
What is homoplasy, and how does it affect phylogenetic tree building?
Homoplasy refers to similarity in traits due to parallel evolution or reversals, rather than shared ancestry. It can mislead phylogenetic tree reconstruction because it creates the appearance of shared traits between unrelated species, leading to inaccurate tree relationships.