A3.2 CLASSIFICATION & CLADISTICS
1/9
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
10 Terms
Outline the classification of organisms and its need
Classification: the organisation & naming of organsims in hierarchical groups called taxa
Needed because of the immense diversity of species
Taxonomy is the classification of living things into groups based on morphological/molecular characteristics
CLASSIFICATION ALLOWS SCIENTISTS TO:
Determine the number of known species
Determine evolutionary relationships: shared common ancestors
Carry out conservation: identifiable species can be conserved
Conduct medical research: identify closely related species with medical benefits
Identify and treat new diseases: COVID-19 classification as a coronavirus
Outline the traditional hierarchy of taxa
king Phillip Came Over For good Sushi!
Kingdom
Phylum
Class
Order
Family
Genus Species
Outline difficulties of classifying organisms in the traditional hierarchy of taxa
Traditional hierarchy does not always correspond to patterns of divergence generated by evolution
Morphological grouping does not always correspond to divergence & shared recent common ancestor (similar traits could be due to convergent evolution)
Taxonomic rank: clear distinction is difficult with hybrids and if a group of organisms needs to be moved from one taxa to another
Species determination: the stage at which two populations are classified as species is difficult to determine/introgression could occur
Outline the clades and its advantages
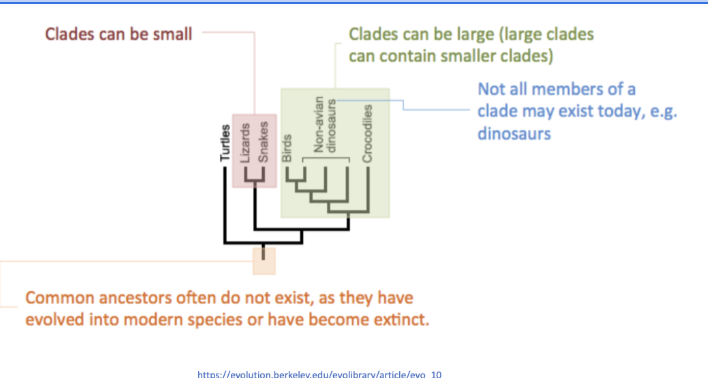
CLADE: Group of organisms that have evolved from a common ancestor
Can be small/nested in larger clades
The common ancestor may be extinct
Organsims in the same clade will share more characteristics/phylogenetic evolutionary relationships
The branch length can indicate the relative number of genetic change
Based on molecular analysis/genetic differences in amino acid sequence/DNA base sequence
predictability suggests evolutionary timelines
Greater differences, the longer period since 2 species had common acnestor
ADVANTAGES:
All the members of a taxonomic group have evolved from a common ancestor
Characteristics are shared within a clade
Characteristics of organisms within the group can be predicted
Outline the use of cladogram
CLADOGRAM:
Tree diagrams that show the most probable sequence of divergence in clades
Shows the same evolutionary relationships & shared characteristics
Organsims in the same clade are more closely related evolutionarily
Nodes represent the splitting of clades from a common acnesotr
Branches at nodes show the time since divergence/separation and the number of differences in DNA
Evidence of placing organisms in the clade:
Base sequence of genes/amino acids of proteins
Morphological traits can be used to assign organisms to clades
Outline the molecular clock
Can be used to estimate the time since divergence
Mutation rates of DNA are steady, but only a probability
Cn gaive estimates
Because mutation rates are affected by the length of the generation time, the size of a population, the intensity of selective pressure and other facotrs
Outline the bases for construction of cladograms
Base sequences of gene & amino acid sequences of proteins is the basis for constructing cladograms:
Biochemical: the study of similar molecules in different species (DNA, RNA, mDNA, hemoglobin, cytochrome C)
Base sequence of a gene, DNA/mDNA — molecular differences can be due to mutations
Amino acid sequences of a protein (hemoglobin, cytochrome C)
Traditional methods use morphology to compare homologous structures, fossils, or comparative anamoly
Outline how cladistics can be used to investigate whether classification of groups corresponds to evolutionary relationships
Classifications of groups may not always reflect evolutionary relationships
Investigations my cladistics can be taken place.
Similar features may be due to convergent evolution (look at difficulties!!)
EXAMPLE: Reclassification of the figwort family
Traditional/Linnaena classification physical similarities were not based on evolutionary relationships
The flower shape & seed capsule evolved through convergent evolution
Different plant species adapted to similar pollinators and similar seed dispersal strategies
DNA evidence (three chloroplast genes) identified different common ancestors
Reclassification of the figwort family was required based on the molecular evidence
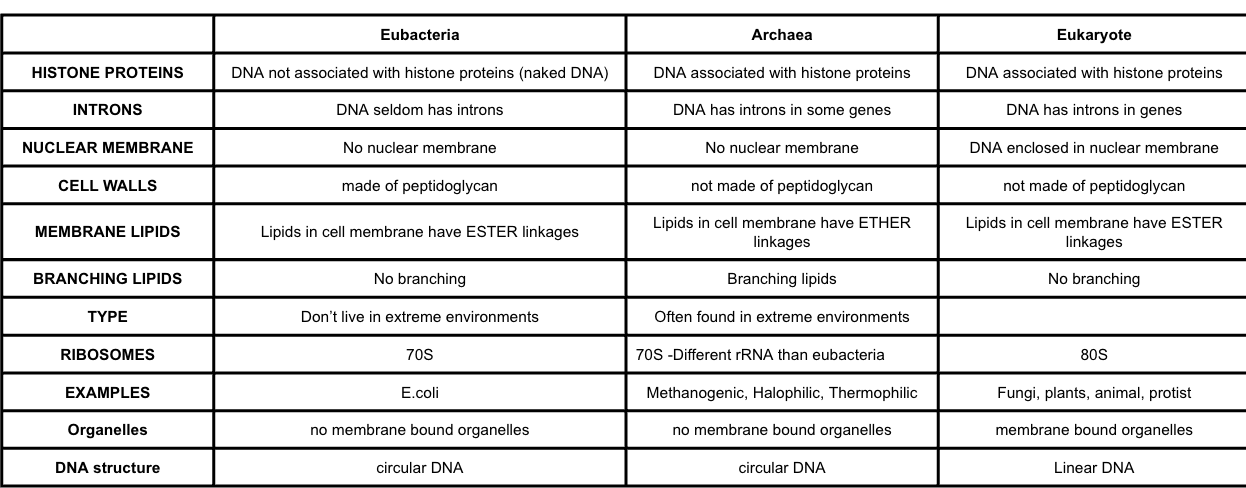
Outline the Classification of all organisms into three domains
All organsims are classified/reclasffieid into three domains using evidence from rRNA base sequences
The three domains are eukarya (eukaryotes), archae (prokaryotes), and eubacteria (prokaryotes)
They share a common ancestor but none of the three domains is an ancestor of the others
Compare & contrast the three domains
Note histone proteins, introns, ribosomes (with rRNA structure), lipids in the membranes