Bacterial Genomics
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
True or false? “All traits of organisms are adaptations”
Not all traits are adaptation.
some are by chance, some are byproduts like the color red in our blood. some are by genetic
drift,some are exception. some was an adaption before and has lost it's function.
some traits can be adaption which help the organism to survive, but not all traits are adaption
True or false? “Individual organisms can evolve during a single lifespan”
While mutations and genetic variations can and do occur within a generation, this is not really
what is referred to when discussing the concept of evolution and to evolve. To evolve one is
observing a longer time distance involving multiple generations, within in a particular
population. One often talk about biological species and not the individual organism.
Phenomena such as positive-
, negative-selection, genetic drift, recombination, HGT and so on
require more than a single individual.Therefore, while it does "seem" like its true, the
statements is far to nuanced and requires circumstances beyond a single individual organism.
True or false? “Genetic drift occurs only in small populations.”
Genetic drift is according to chance, and is occuring constantly in small and large
populations. Genetic drift is more visible in small population. The two main theories for genetic
shift occuring is when you have the bottleneck effect and the founder effect.For both models, it
is by chance that these allels become common and not due to any selection pressure. and
even though the bottleneck and founder effect start off with small populations they become
large populations over time. Viruses transmitted by mosquitoes are often gived as an example
for genetic drift.
What are bacterial genomes constituted of? How are genes divided between them and what is the size difference? What is a chromid?
Chromosomes and plasmids, both of which can be circular or linear.
Differences: Plasmids tend to carry non-essential genes while ribosomal RNA genes are only on chromosomes. They replicate with different mechanisma and are different sizes. The largest plasmids (mega plasmids) are >100kb while the smalles chromosomes (minichromosomes) are 133kb<.
Chromid = plasmid-type replication + plasmid-type maintenance + essential genes.
How big are genomes in the different domains? How is the coding density?
Domain sizes: Eukaryotes > Prokaryotes > Viruses
Eukaryotes are aprox 10e4 to 10e11
Prokaryotes are aprox 10e5 to 10e7
Viruses are aprox 10e2 to 10e6
Coding density = genome size vs nr of proteins
Coding density = viruses > prokaryotes > eukaryotes
Viruses and prokaryotes follow a linear relationship of 1000bp/protein between genome size vs nr of proteins while that of eukaryotes is more curved and plateaued.
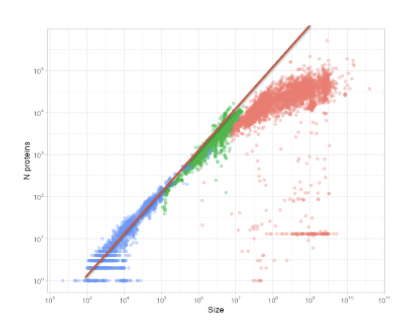
What is the main factor organizing bacterial chromosomes?
The properties of bacterial genome replication strongly affect gene order and placement. The 4 properties of bacterial replication:
Bidirectional replication
Semi-conservative replication = each DNA genome has one new DNA strand and one inherited DNA strand
Asymmetrically replicating strands = leading strand and lagging strand
Replisome moved from origin to terminal
What causes X-patterns on the X-plots?
Symmetrical inversion around the region of origin/terminus and recombination between replication forks because of the close proximity during replication. These are often counter-selective recombinations e.g what was on the lagging strand moves to the leading strand.
What influences gene order conservation between species?
Gene order can be conserved by a neutralist or selectionist approach.
Neutralist = a lack of repeats or a lack of recombinaition machinery
Selectionist = constraints on the gene organization like important co-transcription
What is the mechanism for transfer of genomic islands (GIs)?
Genomic islands (GIs) are regions of a genome prone to HGT carried by phages, plasmids or ICE (integrative conjugative elements).
AKA the machanism of transfer:
Conjugation (physical contact between bacteria using a pilus) - ICE & plasmids
Transduction (delivery of DNA through a phage) - phages
Transformation (uptake from free DNA in the environment) - plasmids
Why do genomic islands become immobilized in genomes?
“GEIs often carry insertion elements or transposons, which may have been implicated in mobilizing genetic material onto or deleting DNA from the element”
What is a pan-genome?
Pan-genome = core genome + auxiliary genome
Core genome = genes shared between all or most members of a taxon
Auxiliary genome = context dependent genes that come from an adaptation based on local competition and environmental pressure e.g mobile elements, GIs, and hypothetical proteins.
What are the theoretical consequences of an open pan-genome?
Closed pan-genome = large core genome + small accessory genomes = dosean’t add new genes often
Open pan-genome = small core genome + large accessory genomes = pan-genome size increases indefinitley when adding new genomes; thus sequencing additional strains will likely yield novel genes.
What does the size of the auxiliary genome depend upon?
Open or closed pan-genome?
Why do conventional typing (serotyping) and core genome phylogenies not match?
The classical and convenient typing of bacteria based on their capsular polysaccharide composition does not reflect the genetic diversity of the species.
Where do HGT integrate and why there?
Integrated elements aren’t usually essential so they make good targets for insertions
Recombination often occurs at the conserved sites flanking homologous sequences
Types of hotspots: scattered, regional hotspots (some genome parts are stable), delocalization (stable chromosomes so hotspots are on plasmids).
What are the molecular mechanisms introducing more GC / more AT in bacterial genomes?
Mutations in the form of selection and drift for example repair enzymes, mutational bias, GC-richness due to stability (selection).
What is (biased) gene conversion? How does it affect GC content?
Gene conversion = one gene allele is replaced by another in a recombination mediated process
Biased gene conversion = many fungi and animals have a conversion bias toward GC nucleotides → more 3rd bases in a nucleotide codon are GC than before. This occurs during recombination or heterozygosity.
What evolutionary forces are at play here?
idk:
The effect of biased gene conversion is greater in species with high variation levels or genomic regions with high levels of recombination.
Prokaryotes also have biased gene conversion with an effect as strong as in the humans genome & in some bacteria.