Haplotypes & Consanguinity
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
Consanguinity
Being descended from the same ancestor as another person.
Consanguineous marriage = matrimony (breeding) between two family members who are second cousins or closer
Globally ____% of children have consanguineous parents
8.5%
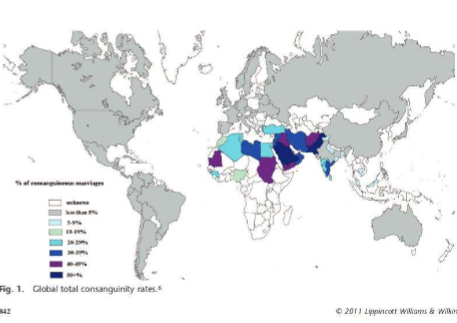
Where is consanguinity seen most in the world
Endogamy is widely practiced across the greater Middle East
Consanguinity is associated with increased risk of what
autosomal recessive disorders
Increases risk of genetic disease from about 2% to about 4% (Probability of having a child without a constitutional congenital defect reduced from 98% to 96%)
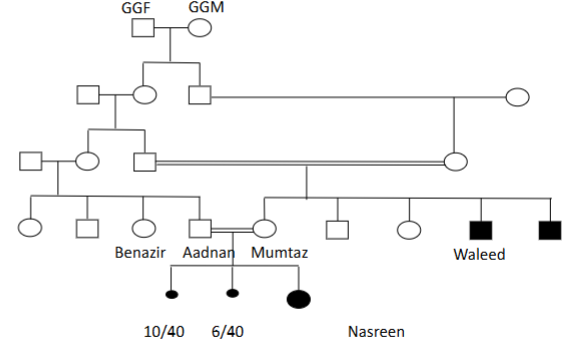
The pedigree suggests what pattern of inheritance ?
Autosomal recessive
Small dots = miscarriage at 10 weeks & 6 weeks
Autosomal recessive deafness accounts for about ___% of congenital deafness
65%
(infection is another cause)
What patterns of inheritance are associated with deafness & what % prevalence are each of them
Recessive (DFNB) – 75-80%
Dominant (DFNA) – 20-25%
X-linked (DFNX) – 1-2%
Matrilineal (mitochondrial) - <1%
What are 2 types of inherited deafness
Syndromic - other symptoms with deafness
Non-syndromic - no other symptoms - just deafness
How many genes are believed to be implicated in deafness
>100
Mutations in what genes can cause hereditary deafness
DFNB1 (13q)
GJB2 (connexin 26 protein)(Cx26)
What mutations of the GJB2 gene are common in Europeans & what is common in Chinese
c.35delG common mutation in Europeans
c.del235C common mutation in Chinese
How is the GJB2 gene checked
Short gene (2 exons) so amplification and sequencing is practical
How does consanguineous mating increase risk for inheriting defecting chromosomes
A bad gene in 1 great grandparent could go to 2 children, if their children / grandchildren have children of their own, they could end up inheriting 2 of the identical bad chromosomes that came from the great grandparent
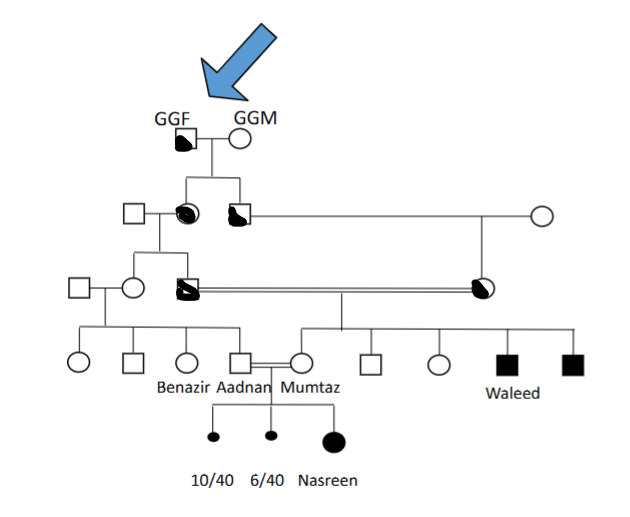
If children all end up deaf from 1 single chromosome from a great grandparent that they have inherited 2 of, does this mean that they have identical relevant chromosomes
All 3 deaf individuals should have practically identical copies of the relevant ancestral chromosome that carries the defective allele
However, crossing over between maternal and paternal means that many offspring do not inherit a full intact version
How do you track Chromosomes or Chunks of Chromosome
Molecular Barcoding
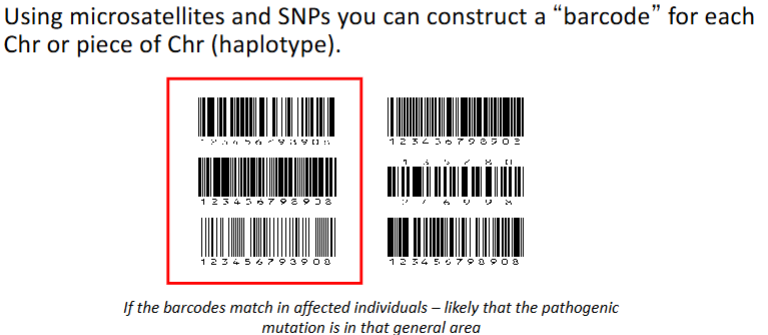
How would you identify deafness-related haplotype by linkage analysis
Perform genomic analysis of polymorphic DNA markers associated with deafness for all family members
See which markers are carried only by deaf members and never by healthy members (segregate with the disorder)
High probability that deafness causing gene is linked to this/these marker(s) (same location on chromosome)
How does Autozygosity mapping work
1. Identify Microsatellite Sequences Associated with deafness loci using dbSNP database
2. Amplify DNA from all of the loci of all family members
3. Determine the size of the PCR products in each individual – affected individuals should all be homozygous for the same haplotype (set of markers)
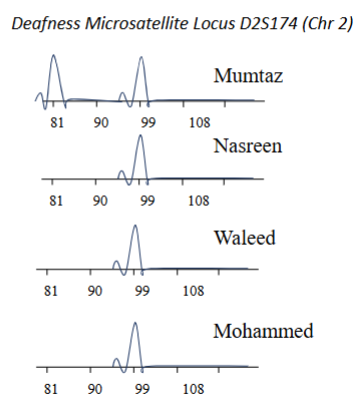
Which of these are homo/heterozygous for the particular haplotype at this locus
Homozygous = Nasreen, Waleed & Mohammed (Single products suggests that they are homozygous)
Mumtaz is heterozygous
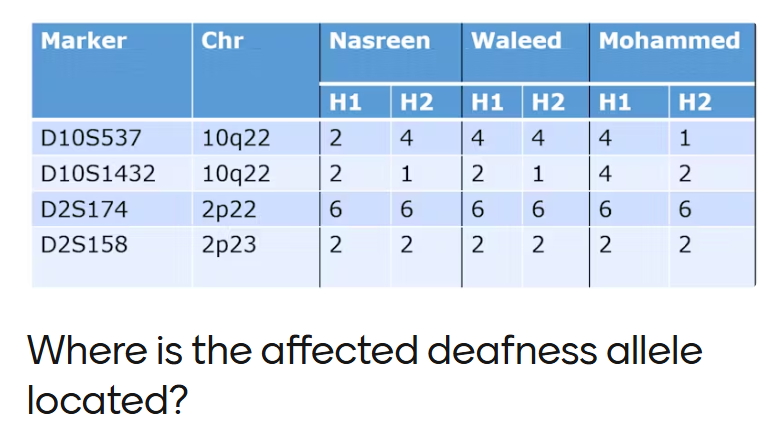
The deafness gene is most likely located on chromosome 2p22–2p23, because both all affected individuals are homozygous for this region; (6,6) and (2,2), consistent with inheritance of a recessive mutation from both parents.
(6,6) = allele number - means H1 and H2 are homozygous at that locus (both copies of the gene are identical)
What is an important assumption not to make when dealing with linkage
Association is not causation - Linkage of a trait or disorder with a gene/DNA sequence does not imply that the gene/DNA sequence associated with the trait is important in the pathogenesis of the condition
Coefficient of Relationship (COR)
The proportion of alleles that 2 people share by virtue of having one or more definable common ancestors
Coefficient of Inbreeding (COI)
The proportion of loci at which the person is expected to be homozygous because of the consanguinity of the parents
or
The probability that at any given locus the person receives two alleles that are identical by descent
How do COI values compare to CORs in fraction form
COI = half the coefficient of relationship of the parents
How can we calculate the COR in outbred families? (what % of genes are shared between:
parents & their children
full siblings
grandparents & their grandchildren
uncle/aunt with nephew/niece
cousins
• Parents share 50% of alleles with their children
• Full siblings share 50%
• Grandparents share 25% with grandchildren
• Uncle/Aunt share 25% with Nephew/Niece also
• Cousins share 12.5%
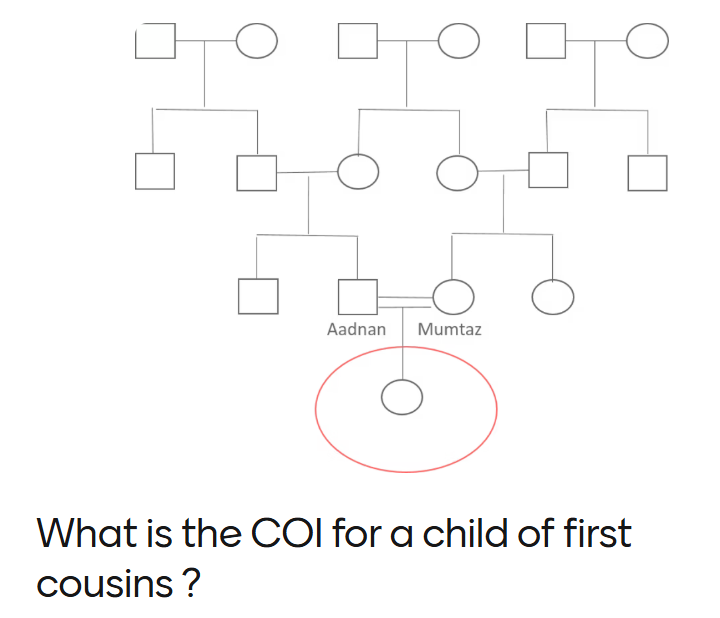
6.25%
Allele Frequency definition
The incidence of a gene variant in a population
Allele Frequency formula

Allele frequencies are a reflection of what in a population
genetic diversity
True/False: allele frequency of 40% means 40% of the population have a copy of the allele
False
If 10% of 100 people are homozygous for allele R1 (20 copies), and 20% are heterozygous and for allele R1 – (20 copies)
Allele frequency is?
40/200 = 20%
The Hardy–Weinberg principle, also known as the Hardy–Weinberg equilibrium, model, theorem, or law, states what
allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences (including selection)
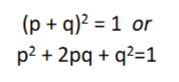
What do p and q mean in the hardy-weinberg equation mean
p = frequency of allele “A”
q = frequency of allele “a”
p + q must = 1 (q = disease frequency, p = non-diseased frequency)
(haPPy & healthy)
Alleles distribute randomly only under certain assumptions. Name 1 such assumption
the absence of selection and random mating (a panmictic population)
(doesn’t actually happen in real life all the time)
When these conditions apply, a population is said to be in Hardy–Weinberg equilibrium
Hardy-Weinberg Equilibrium: In clinical genetics it is mainly used for what
to determine the frequency of heterozygosity (carrier frequency) in autosomal recessive heritable disorders where only the disease frequency (q2) is known
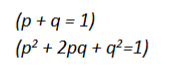
Allele frequency (p) for R1 = 40% (0.4)
What proportion of the population have allele R1?
