mod 4 cont'd: genome 2,3,4
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
54 Terms
what is the folding of RNA?
complex 3D structures
stem + loop(, G-U stabilizes end of hairpin), hairpin structures
what does folding enable in RNA structure?
catalytic and structural roles.
what are all the types of RNA?
mRNA, tRNA, rRNA, snRNA, snoRNA, miRNA, siRNA, IncRNA
what is RNA polymerase?
synthesis of complementary RNA, reads 3’→5’ but synthesies in 5’→3’
is primer required for initiation of RNA synthesis?
no primers are required
what are promoters?
DNA sequences upstream of a genes transcription start site +1
the TATA box is the recognition site. serves as the start codon
what does the TATA box define?
transcription start site and direction
what are the three steps of transcription?
initiation: RNA polymerase binds to promter, unwinds DNA, begins RNA synthesis
elongation: RNA polymerase moves along DNA synthesizing RNA
termination: RNA polymerase encounters trancsription stop signal in DNA
how does bacterial promoters start transcription?
-35 element
upstream from +1; recognition and binding site for sigma factor ( enables promoter recognition and transcription initiation)
-10 element
A-T rich region that melts easily to form open complex to get transcribed.
how does elongation occur in prokaryotic transcription? (bacterial)
after 10nts sigma factor dissasociates and elongation complex forms
RNA polymerase moves along template, synthesizing RNA 5’→3’
what are the three different RNA polymerases?
pol. 1 most rRNA
pol.2 mRNA
pol.3 tRNA
multi subunits, RNA transcript exits via channel
what does the eukaryotic transcription process require?
general transcription factros
always needed to initiate
what is the preinitiation complex?
GTF assemble with pol.II to form the preinitiation complex (PIC)
corelelments contain the TATA box, BRE, initiator
how does initiation start from promoters?
TFIID
TBP( TATA binding protein)subunit binds to TATA box
TBP associated factors (TAFs) needed for binding to occur
recruitment of GTF’s
TFIIB positions RNAPII
TFIIA
recruitment of RNAPII-TFIIF
TFIIF stabilizes interaction between RNAPII and TBP & TFIIB
recruitment of additional GTF’s
TFIIH kinase and helicase activity
TFIIE
PIC is now formed and RNAPII is active
where are mRNA’s processed?
in the nucleus
transported out to the cytosol for translation to take place
how is nucler transport of mRNA’s facilitated?
through nuclear pores
before processing can occur RNA processing steps must occur first
(before= pre-mRNA)
what are the steps of RNA processing?
capping- modification of 5’ end and takes place after 25 nts. includes guanine + methyl groups
splicing- removes introns, joins exons
polyadenylation- added poly A tail at 3’ end for degradation during transport
what are the structures of mRNA’s?
continuous coding sequence
5’ methylated cap
3’ poly-A tail
what are split genes?
difference in in size between hetereogenous nuclear DNA
intervening sequences = intron (noncoding)
expressed sequences = exon (coding)
why does RNA splicing increase protein diversity?
different combinations of exons joined making unique mRNA’s
→ enables tissue specific protein expression
what is a spliceosome?
removes introns, joins exons
what is transcriptional control regulated by?
general TF’s
bind to core promoter, recruits RNA pol. II
sequence specific TF’s
bind to various regulatory sites of particular genes
what are transcriptional activators?
stimulate transcription
what are transcriptional repressors?
inhibit transcription
what two domains do TF’s contain in structure?
DNA binding domain: region specific DNA motifs
activation domain: interacts w/ other proteins to modulate transcription
they form dimers for stability and specificity
what kind of roles do TF’s play in gene expression?
extent of transcription depends on combination of TF’s bound to upstream regulating elements
combinations can differ in cells of different type tissue and stages
5-10% of genes encode TF’s
**combinatorial control of transcription: Oct4
what is combinatorial control of transcription Oct4?
TF for its own synthesis that matches the downstream factor
what are embryonic stem cells?
indefinite self-renewal, pluripotentcy
controlled by for TF network
these re-expressing factros reprogram adult cells inducing pluripotent stem cells
where does RNA splicing start?
5’ GU start
3’AG end
what is the sliceosome composed of?
small nuclear RNA’s (snRNA and associated proteins snRNP’s)
what do the snRNPs recognize?
they recognize splice sites at intron exon boundaries (GU-AG rule)
this assembles a step-wise process on pre-mRNA bringing exons together
catalyzes two transferication rrxn intron releases and exons joined
dynamic complex components are recycled for multiple splicing events
translation
what are ribosomal proteins?
synthesizes in the cytoplasm and imported into the nucleus
RNA genes are clustered i the nucleolus and transcribed by RNA pol.I
what does the nucleolus act as?
acts as the cells ribosome factory
what is the structure of the euk. ribosome?
4 distinct rRNA’s
2 subunits
small-18s
large- 28s, 5.8s, 5s
where are the five “cut” locations of RNA processing?
1 and or 5
2 or 5?
what are the nucleotide modifications?
methylation
conversion or uridine→pseudouridine
preformers by snoRNPs
what are the properties of the genetic code?
redundant
conservative
unambiguous
universal
what is the start codon?
AUG
(methyionine)
what are the stop codons?
UAA, UAG, UGA
what is the role of tRNA’s?
decoding the codon
they match mRNA codon w/ amino acid it codes for
each linked to a specific amino acid
recognizes the mRNA via the anticodon
what is the unique structure of tRNA’s and why is it shapped like that?
complimentary intrachain base pairing
cloverleaf
anticodon loop:pairs w/ codon in mRNA
AA acceptor arm 3” CCA sequence: binds the amino acid
what is the wobbble hypothesis?
how multiple codons code for single amino acid
interchangibility of base in 3rd position
how does the tRNA recognize amino acids?
depends on aminoacyl-tRNA-synthatases that are unique to every amino acid
they covalenty link amino acids to tRNA 3’ end
what is initiator tRNA?
newly made proteins all have methionine as first a.a. at N-terminal (that is later removed in post translation modification)
→the only amino acid that can bind to P site when large ribosomal subunit is not present
what is tRNA charging?
aminoacyl-tRNA-synthetases attach the correct amino acid to its tRNA
**tRNA’s translate codons into amino acids
accuracy depedns on proper charging and anticodon pairing
initator tRNA sets reading frame for translation
give an overview of translation
initiation: ribosome assembles on mRNA and finds the start codon
elongation: amino acids are added as ribosomes moves along mRNA
termination: stop codon reached, completed polypeptide is released
**requires GTP and protein factors
**occurs on ribosomes in the cytoplasm
how does translation in bacteria differ from euk.?
no 5’ cap
only 3 initiation factors
no scanning step (assembles directly at start codon)
what does the info stored in the mRNA determine?
the sequence of aminoacyl-tRNAs that the ribosome accepts
what are the binding sites on the ribosome?
A(aminoacyl): binds incoming aminoacyl-tRNA carrying new a.a.
P(peptidyl): holds the tRNA w/ the polypeptide
E(exit): release empty tRNA
explain the steps of the synthesis from the ribosome
the second aminoacyl-tRNA binds to A site
GTP hydrolysis releases EF-TU or eEFIA positioning to new A site
catalyzed by peptidyl transferase
growing peptide is transferred from P-site tRNA to a.a. on the A-site
—after bond formation—
translocation
binding of an elongation factor and GTP hydrolysis
ribosome shifts nucleotides (one codon) in the 5’ →3’ direction
release of deacylated tRNA
leaves the ribosome, emptying the E-site
what are the steps of termination?
occurs at stop codon
requires release factors → recognizes stop codons
alter the ribosomal peptidyl transferases
dissasociation of the mRNA from the ribosome, diassembly of the ribosmes
what does quality control take place?
nonsense-mediated decay (NMD)
mRNA surveillance pathway that detcts transcription with premature stop codons
degrades faulty mRNA’s to prevent production of shortened nonfunctional proteins
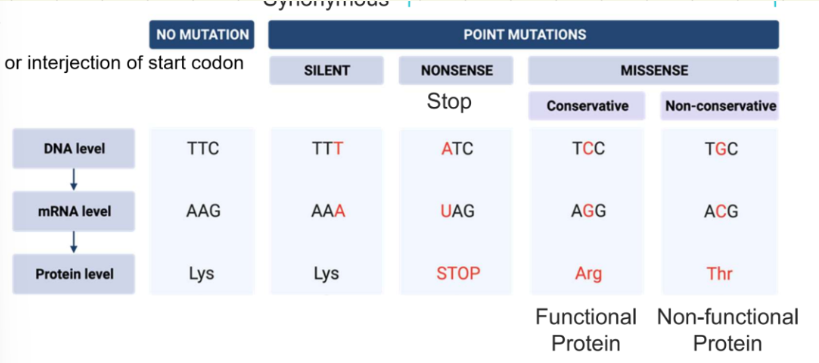
different mutations