D1.1 DNA REPLICATION
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
11 Terms
state why DNA replication is essential
it is important and required for reproduction, growth and tissue replacement in multicellular organisms
state when DNA replication occurs
occurs during S phase of interphase, in preparation for mitosis
state the directionality of DNA replication
DNA polymerase adds new nucleotides joinind from 5’ to 3’ end of the existing chain
synthesised in 5’ to 3’ direction
explain the process of DNA replication
helicase occurs at the origin of replication and unwinds the double helix, separating the two strands of DNA by breaking the hydrogen bonds between the complementary bases and creating a replication fork so each strand can act as parent strands (template strands) for replication (5’-3’ leading strand, 3’-5’ lagging strand).
DNA gyrase helps reduce the torsional strain created by the unwinding of DNA
single-stranded binding proteins (SSBs) bind to separated strands to prevent them from re-annealing or forming secondary structures.
DNA primase generates a starting point by synthesizing a short RNA primer complementary to the DNA template strand (serves as initiation point for DNA polymerase III)
DNA polymerase III adds the nucleotides in the 5’ to 3’ direction according to complementary base pairing, continuous on the leading strand, discontinuous on the lagging strand
DNA polymerase I removes RNA primers and replaces them with DNA nucleotides
DNA ligase joins okazaki fragments together on lagging strand to form a continuous stand by covalently forming phosphodiester bonds
compare leading vs lagging strand
leading strand: DNA polymerase III moves towards replication fork (copies continuously) (replication has to be initiated with RNA primer only once on the leading strand)
lagging strand: DNA polymerase III moves away from replication fork (copies in fragments, discontinuousky) (copied in series of short fragments) (replication has to be initiated with RNA primer repeatedly on the lagging strand)
explain what DNA proofreading is
fixes errors which result in mutation
DNA polymerase III proofreads the added nitrogenous base, removing the nucleotide if the base paring is wrong by breaking the hydrogen bonds
exonucleases remove the sequence surrounding base if mismatch is undetected
outline the process of polymerase chain reaction amplifying DNA
amplify large quantities of DNA from small samples
denaturation: heat the sample to around 90°C, which will cause the double stranded DNA to denature and separate into two single strands
annealing: the sample is left alone until temperature is lowered to around 55°C, allowing the primers within the sample to anneal to their complementary sequences on the single stranded DNA templates, which serves as a starting point for DNA synthesis
elongation: the temperature of the sample is heated to around 75°C, which is the optimal temperature for taq polymerase to function and synthesize new DNA strands complementary to the template strands as it is isolated from heat-resistant bacteria
outline the process of gel electrophoresis seperating DNA
seperate DNA fragments based on mass/size
the DNA is cut into small fragments by restriction enzymes
the sample is then placed in a block of gel in the electrophoresis chamber where the positive electrode is positioned towards the end of the gel
as the electrophoresis chamber is turned on, this will apply an electric current to the gel
since DNA has a (-) charge (phosphate backbone), it will be attracted to the oppositely charged (+) end of the electrode, migrating the gel towards the other end
smaller samples are less impeded by gel, they move faster and farther compared to larger samples
state how PCR and gel electrophoresis might be applicated in real life
DNA profiling: identifying individuals by DNA profiles
collect sample and amplify using PCR
create fragments through restriction enzymes
seperate using gel electrophoresis and compare
high # of markers, low probability of false match
paternity testing: all fragments produced by mother + father
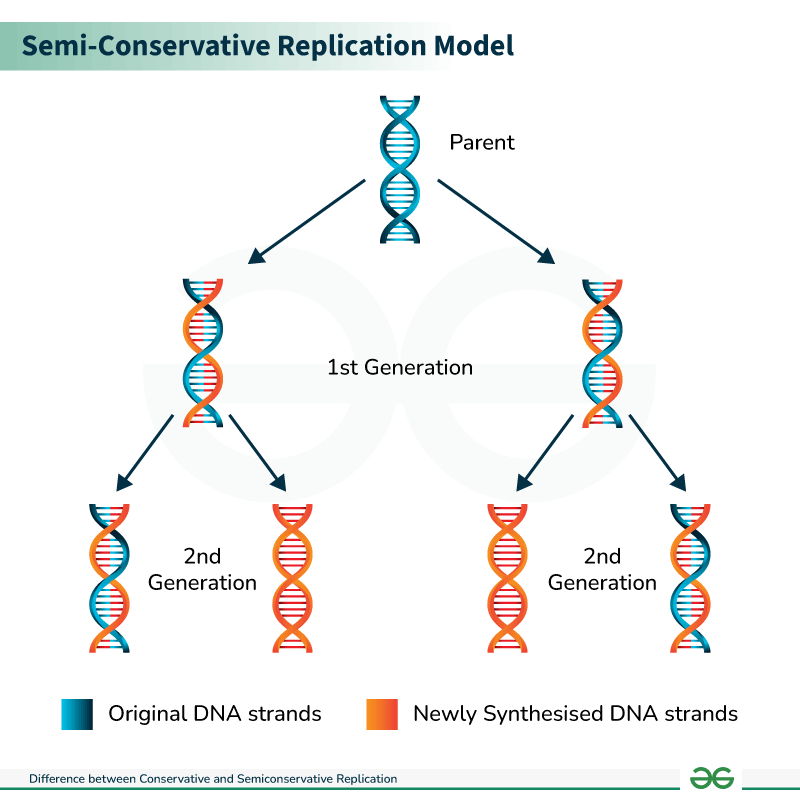
explain the semi-conservative nature of DNA replication and role of complementary base pairing
each DNA copy contains one old strand and one new strand
pre-existing strands act as templates for newly synthesised strands
template strand (conserved) + newly synthesised stand (x consereved)
each nitrogenous base only pairs with one complementary partner which allows in a high degree of accuracy
A-T: 2 hydrogen bonds
C-G: 3 hydrogen bonds
outline how the hershey and chase experiment provided DNA as genetic material
experiment is meant to determine wether DNA or protein is the genetic material
2 batches of sample viruses composed of DNA and protein were created for P32 (to trace DNA) and S35 (to trace proteins) radioisotopes to infect the bacteria
after mixing process and letting it sit etc, P32 isotope was found inside the bacterial pellet after centrifugation indicating that the bacteria entered the DNA
S35 isotope was found on the supernatant after centrifugation indicating that the bacteria did not enter the protein
2 separate batches of T2 (simple virus composed of DNA and protein) phages were created for both radioactive isotopes (batch 1 - labelled with ³²P to trace DNA, batch 2 - labelled with ³⁵S to trace protein) to be infect the bacteria
after allowing time for infection, the mixture of bacteria and phages was blended to separate the phage coats from the bacteria
Basically the ³²P radioisotope was found inside the bacterial pellet after centrifugation therefore indicating that the DNA entered the bacteria
As for the ³⁵S isotope, the radioactivity remained supernatant after centrifugation meaning that the protein did not enter the bacteria