Citric acid cycle & ETC
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
what are the two main functions of Coenzyme A?
1) activation of the acyl groups for transfer by nucleophilic attack
2) activation of the alpha-hydrogen of the acyl group for abstraction as a proton
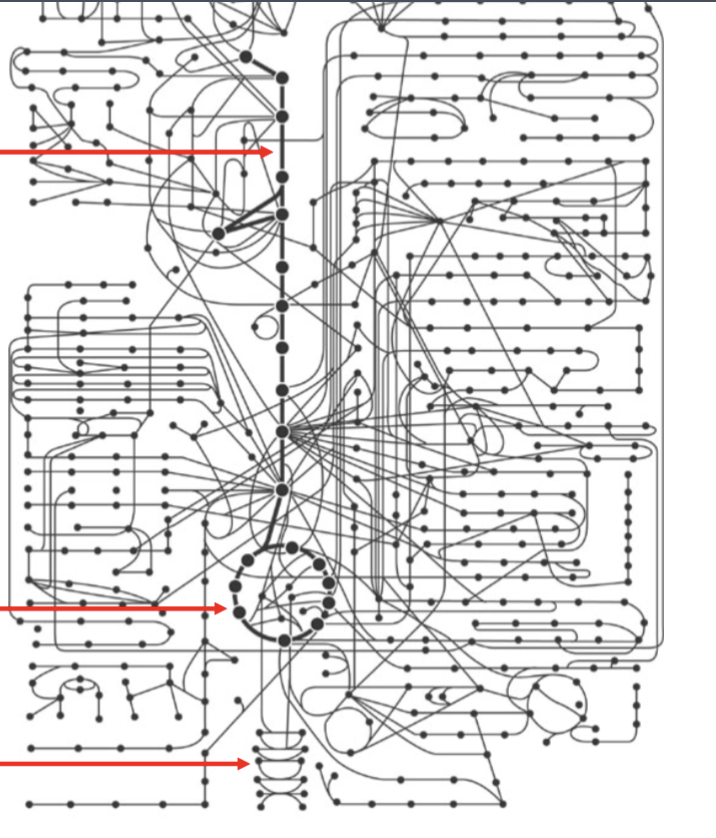
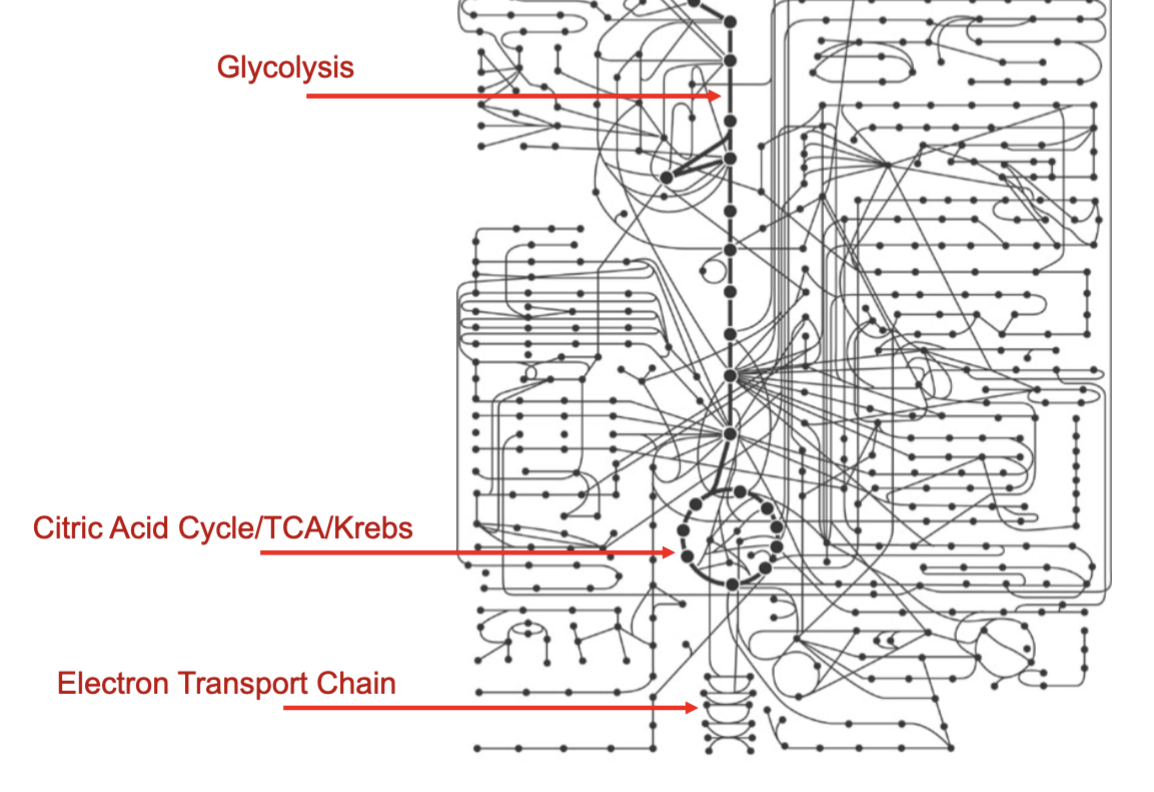
Identify these three points in the metabolic map
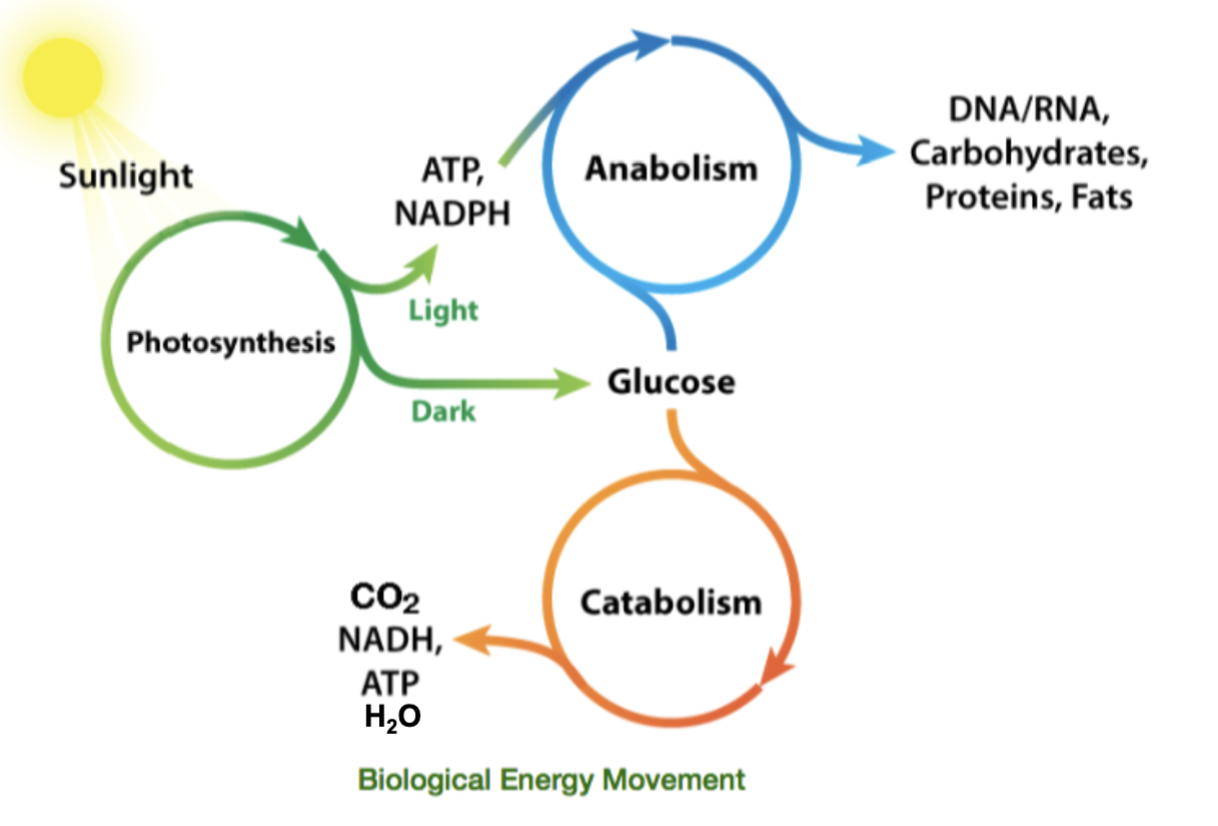
biological energy movement starting from sunlight to photosynthesis
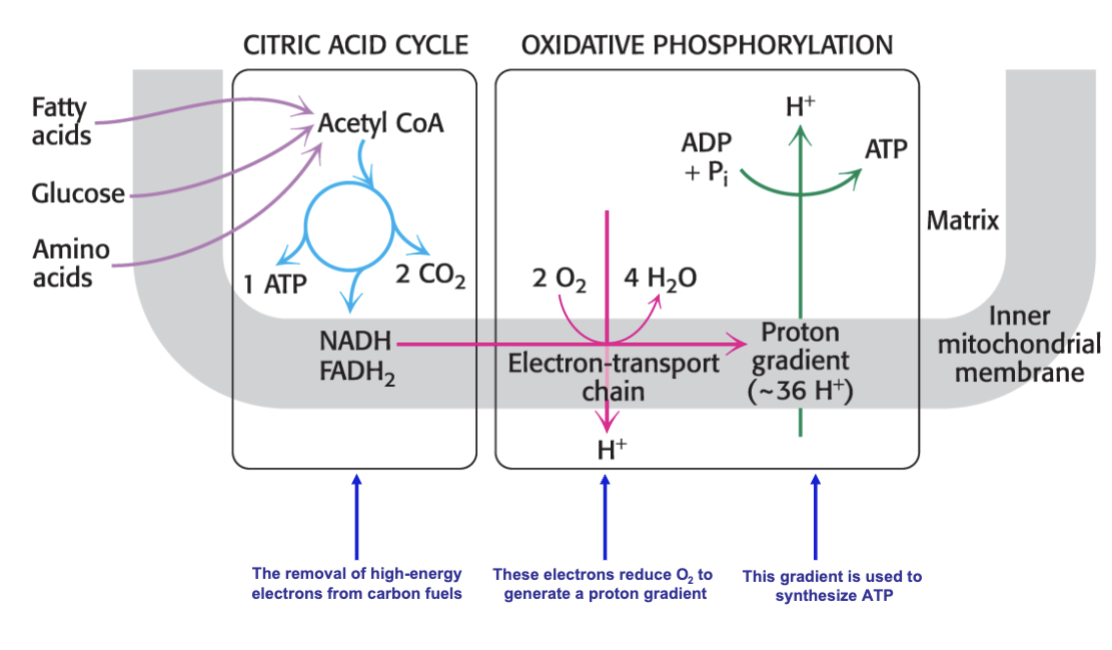
What is the central role of the citric acid cycle?
to generate reducing equivalents:
OXIDIZE acetyl-CoA to generate ATP (energy) and NADH / FADH2 (high energy electron carrieres)
Other than glucose — glycolysis —> pyruvate, what other molecules can become acetyl-CoA for the TCA cycle
Fatty acids and amino acids
in cellular respiration, where is the site of citric acid cycle vs. oxidative phosphorylation
inside mitochondrial matrix for TCA and through membrane for oxidative phosphorylation
name the enzyme classes in order starting from citrate
Is Orange Orange Truly Orange LOL
isomerase
oxidoreductase
oxidoreductase
transferase
oxidoreductase
lyase
oxidoreductase
lyase
—> first step starts at 8
What are the products of oxidizing one Acetyl CoA?
ATP, 2CO2, 8e-
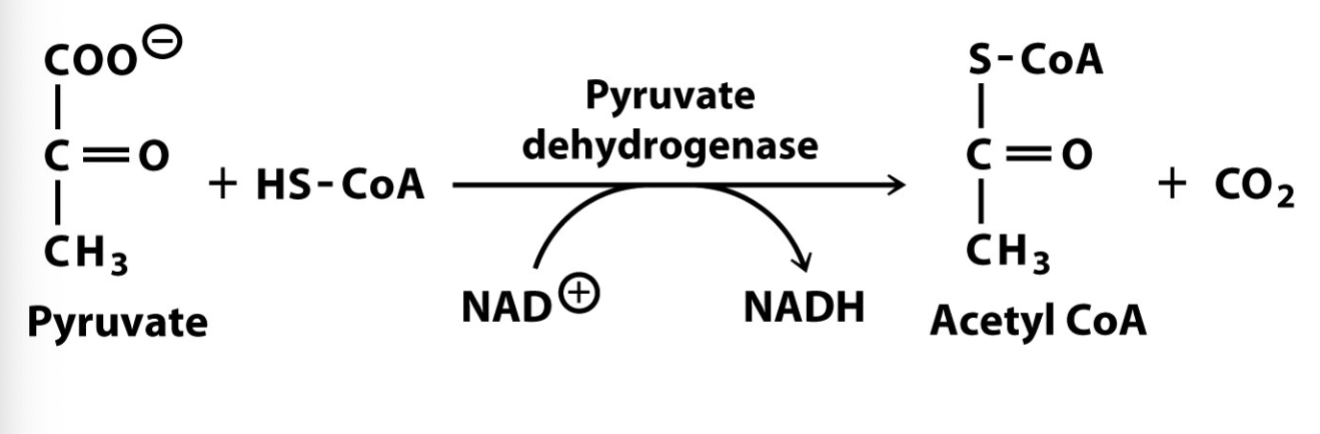
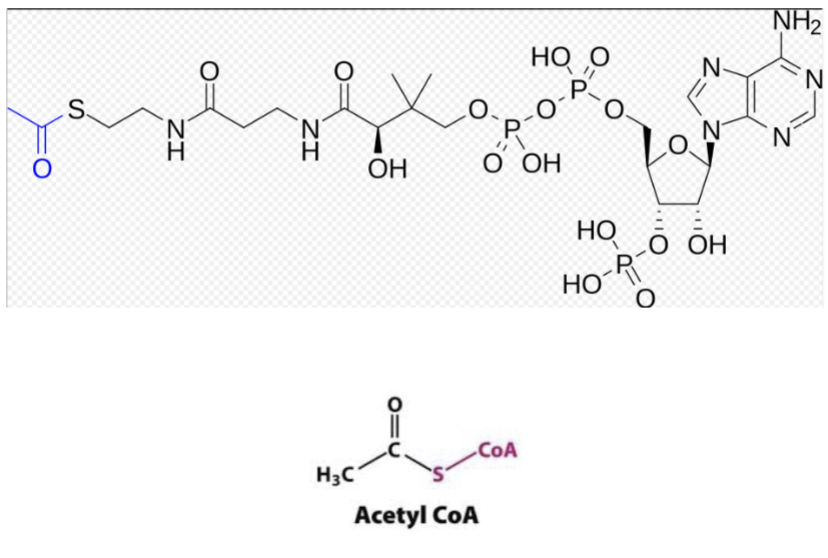
chemical structure of acetyl CoA?
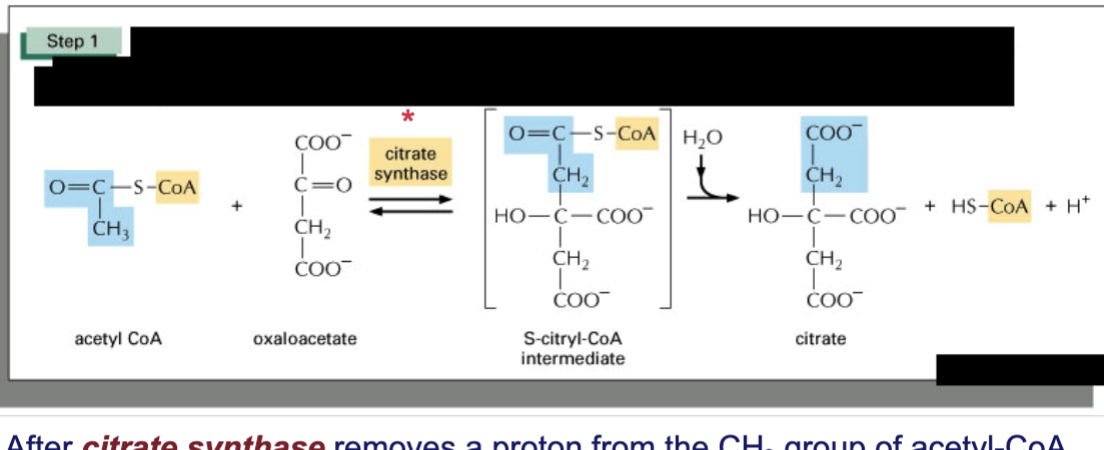
Describe the first step of TCA cycle & spontaneity
big negative delta G
acetyl Co-A is condensated with oxaloacetate to form citrate
control point
catalyzed by lyase citrate synthase, an allosteric enzyme that is inhibited by NADH, ATP, and succinyl-CoA
citrate synthase removes proton from CH3 group of acetyl-CoA —> negative charge forms bond to carbonyl carbon of oxaloacetate
reaction driven strongly forward by hydrolysis of CoA
Describe the second step of the TCA cycle & spontaneity
positive delta G
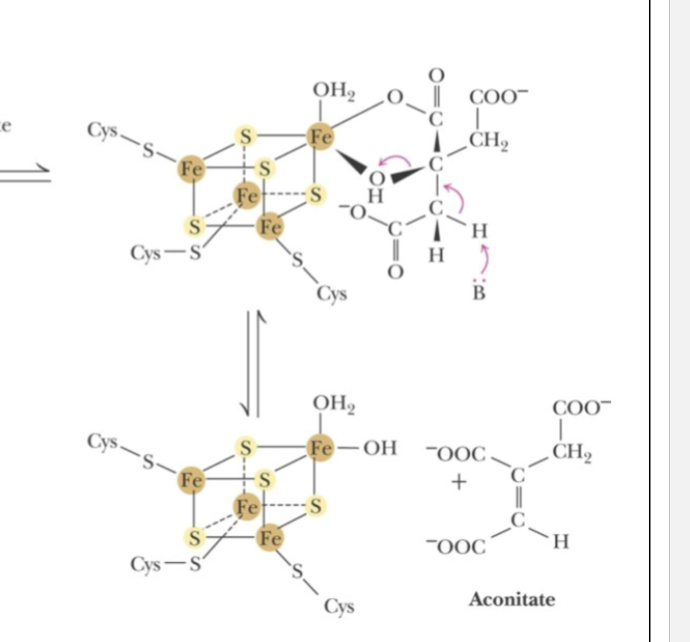
Isomerase (aconitase, iron-sulfur cluster) catalyzes reaction where OH group (as water) is removed from citrate and then added to the neighbouring carbon.
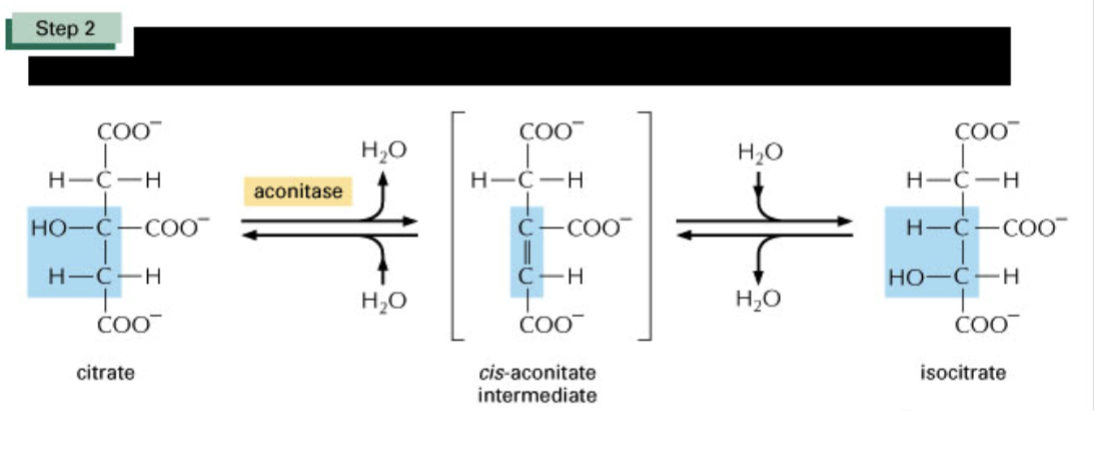
aconitase:
Fe2+ binds to vacant position of the cluster —> activates catalyst
added iron atom coordinates the citrate C-3 carbonxyl and hydroxyl groups and acts as lewis acid, accepting electron pair from hydroxyl group making it better leaving group
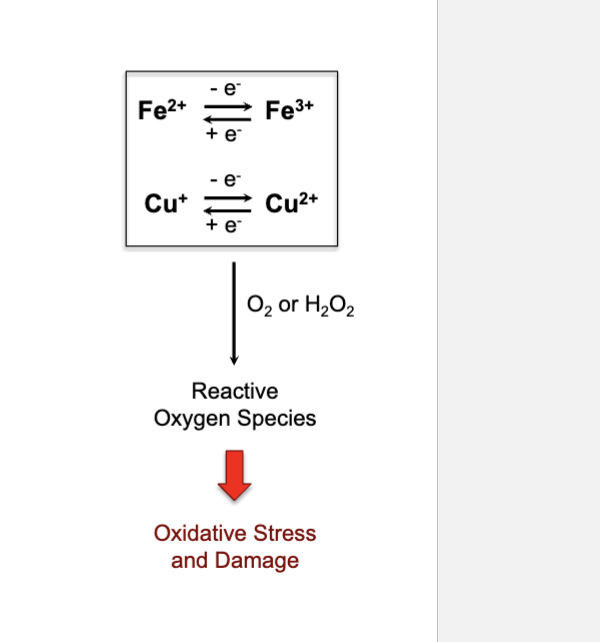
How and why are redox-active metals tightly regulated in the cell?
regulated by labile pools, enzymes, chaperones, storage, and subcellular organelles
can cause oxidative stress:
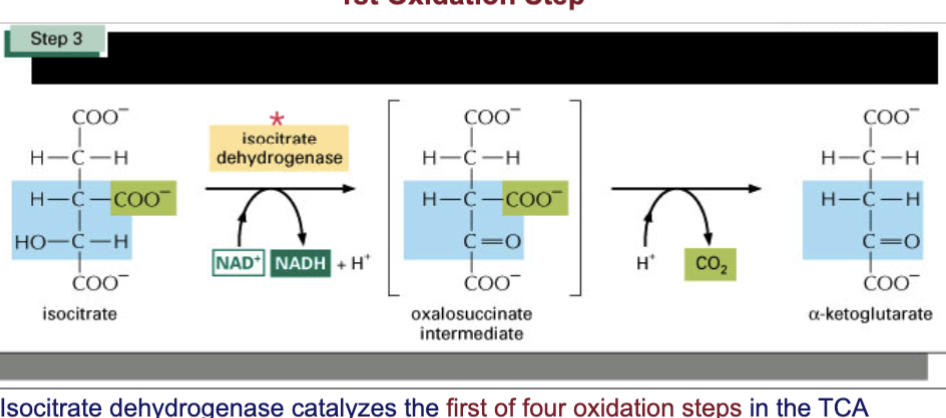
Describe step 3 of TCA cycle & spontaneity
small negative delta G
control point in the cycle: linked to ET chain by NADH production
the first (1/4) oxidation steps in the cycle
carbon of isocitrate w/ the OH group is converted to a carbonyl group with oxidoreductase
NAD —> NADH + H+
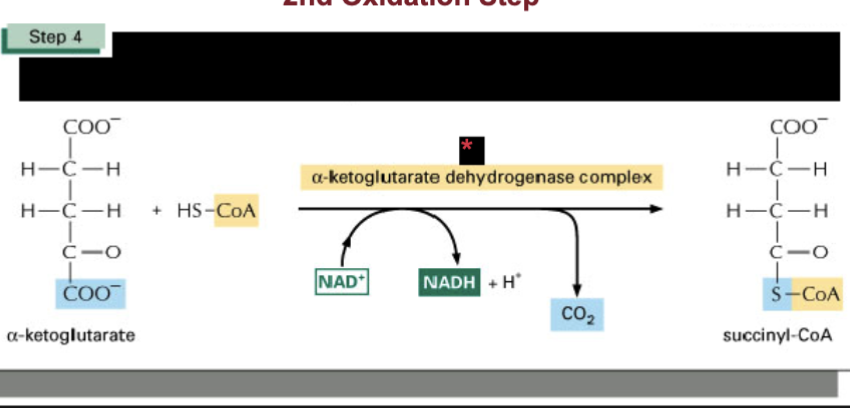
Describe step 4 of TCA cycle & spontaneity
small negative delta G
control point in the cycle
2nd (2/4) oxidation step in the cycle
oxidoreductase catalyzes oxidation that makes NAD+ —> NADH + H+ and CO2
produces high energy thioester bond to CoA
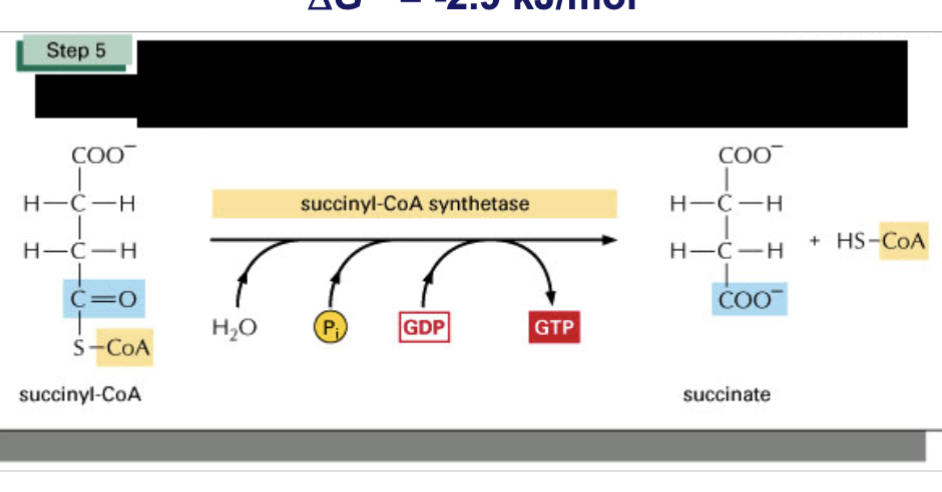
Describe step 5 of TCA cycle & spontaneity
transferase catalyzes phosphate molecule from solution displacing CoA, forming high energy bond to succinate
this phosphate is passed from GDP —> GTP (in plants/bacteria, ATP is formed instead)
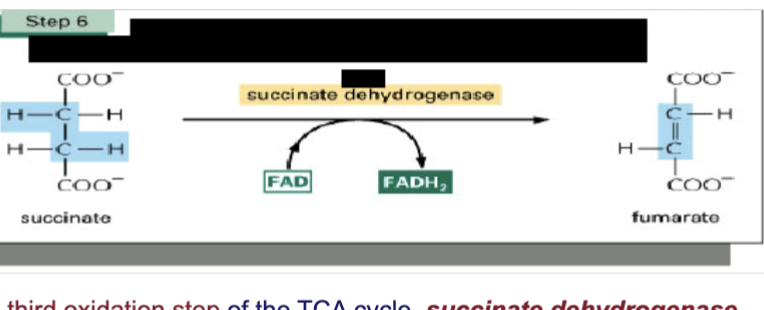
Describe step 6 of TCA cycle & spontaneity
delta G is 0 kJ/mol
3rd (3/4) oxidation step in the TCA cycle
oxidoreductase (succinate dehydrogenase, in eukaryotes —> only enzyme in cycle that is membrane bound) catalyzes reaction where FAD removes two H atoms from succinate to form fumarate (double bond)
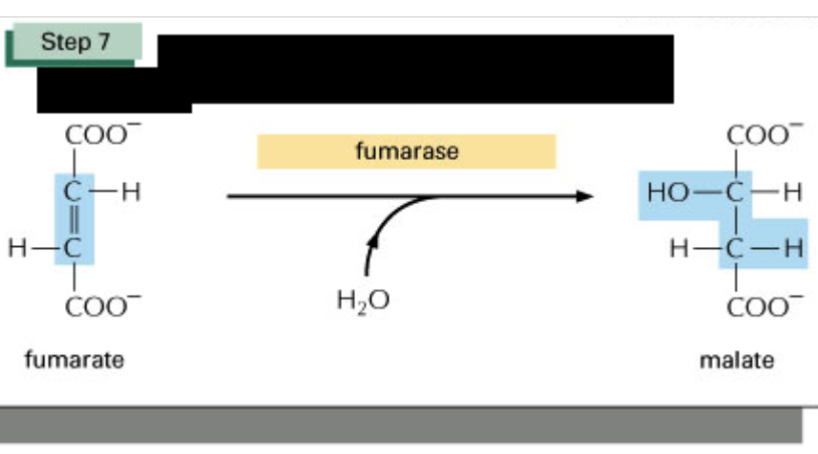
Describe step 7 of TCA cycle & spontaneity
small negative Delta G
Lyase (fumarase aka fumarate hydratase) + water to place hydroxyl group
Describe step 8 of TCA cycle & spontaneity
Delta G positive, but in cells oxaloacetate is continually removed by exergonic citrate synthase (step 1) ** coupling of reaction!
4th (4/4) oxidation step
last reaction of TCA cycle
oxidoreductase (L-malate dehydrogenase) catalyzes oxidation of malate to oxaloacetate
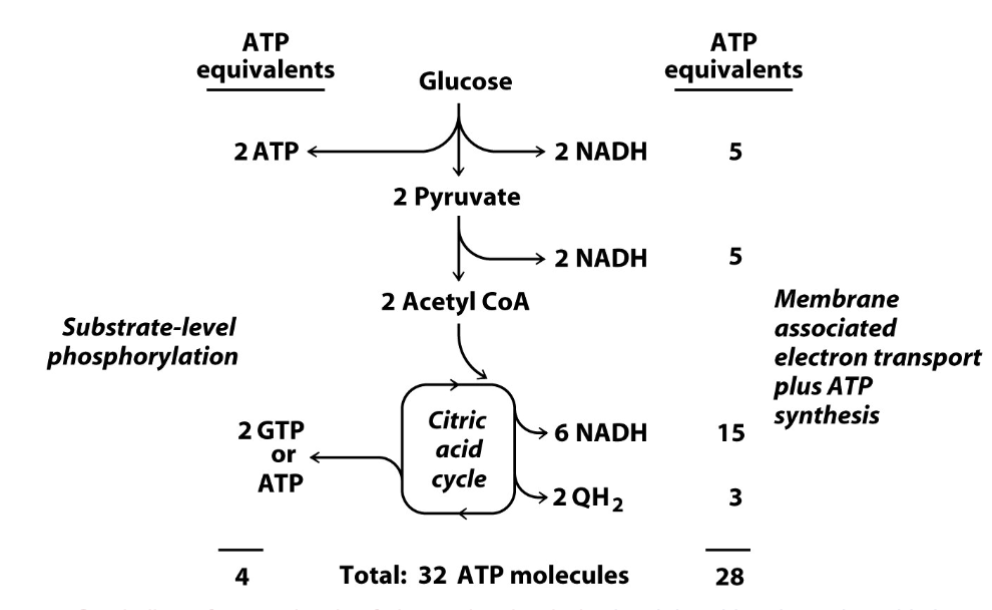
total number of ATP produced by the catabolism of one molecule of glucose by glycolysis, the citric acid cycle, and reoxidation of NADH and QH2
32
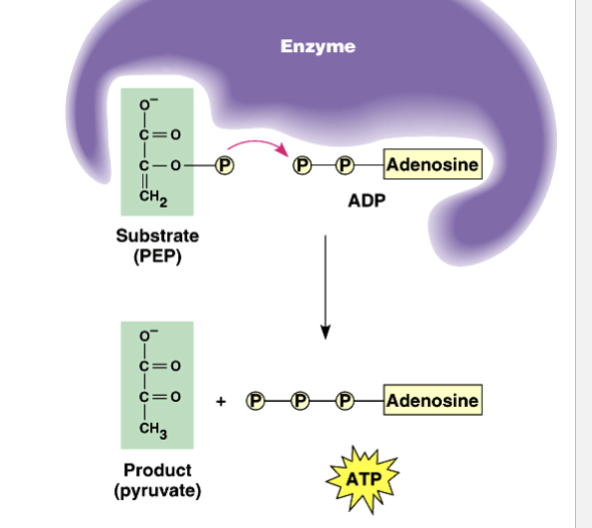
What happens during substrate level phosphorylation, when does it occur?
enzyme transfers a phosphate group from substrate (PEP) molecule to ADP —> ATP
the PEP is derived from the catabolism of glucose
substrate level phosphorylation occurs during glycolysis and in TCA cycle
TCA cycle provides intermediates for biosynthesis, give three examples
alpha-ketoglutarate transaminated to make glutamate —> make purine nucleotides
Succinyl-CoA can be used to make porphyrins
fumarate and oxaloacetate can be used to make several amino acids and pyrimidine nucleotides
The electron transport chain is a series of _______ _______ reactions that transfer electrons from ______ and ______ to ______
The electron transport chain is a series of coupled redox reactions that transfer electrons from NADH and FADH2 to oxygen
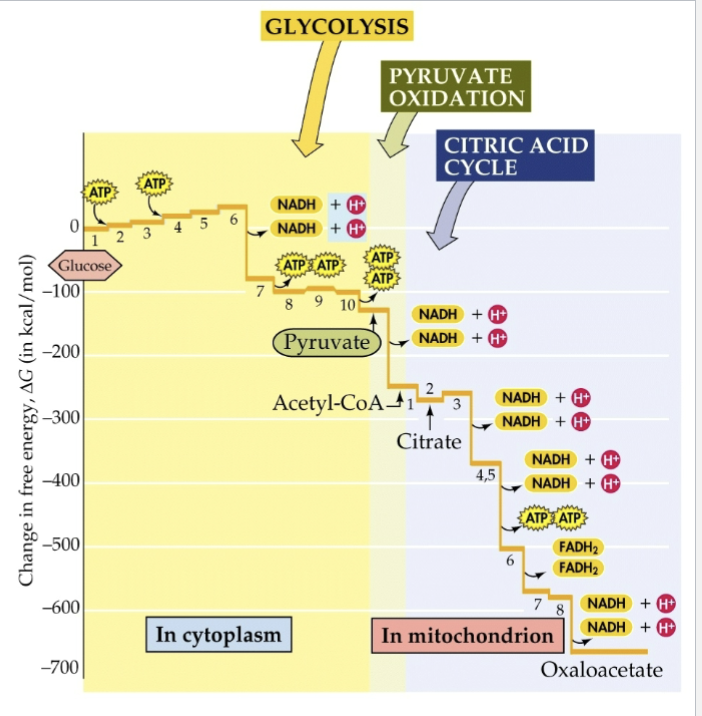
Describe the change in free energy by glycolysis and the TCA cycle
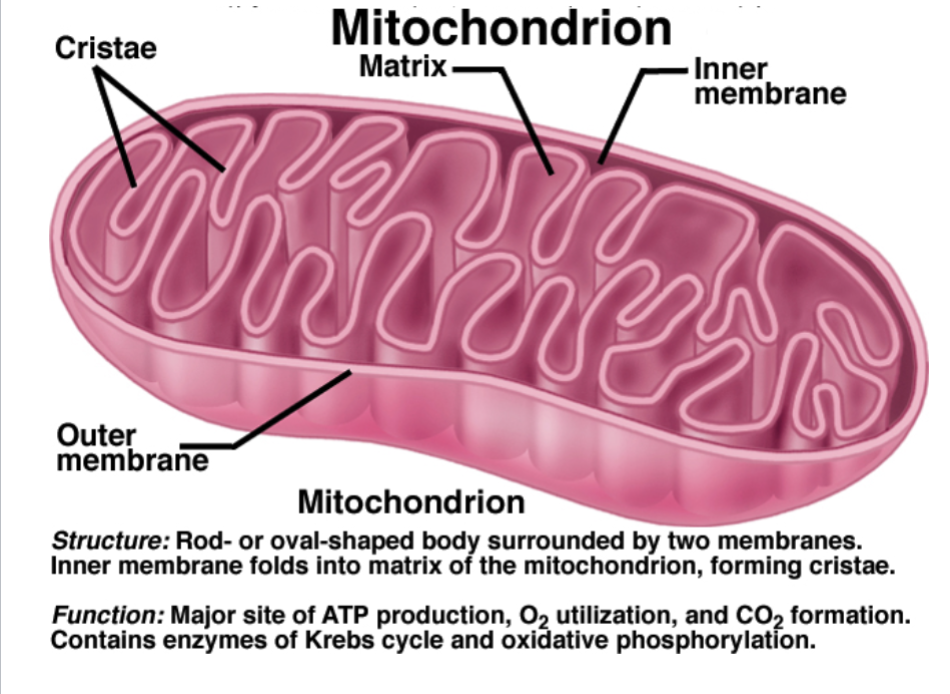
Describe the structure and function of mitochondrion
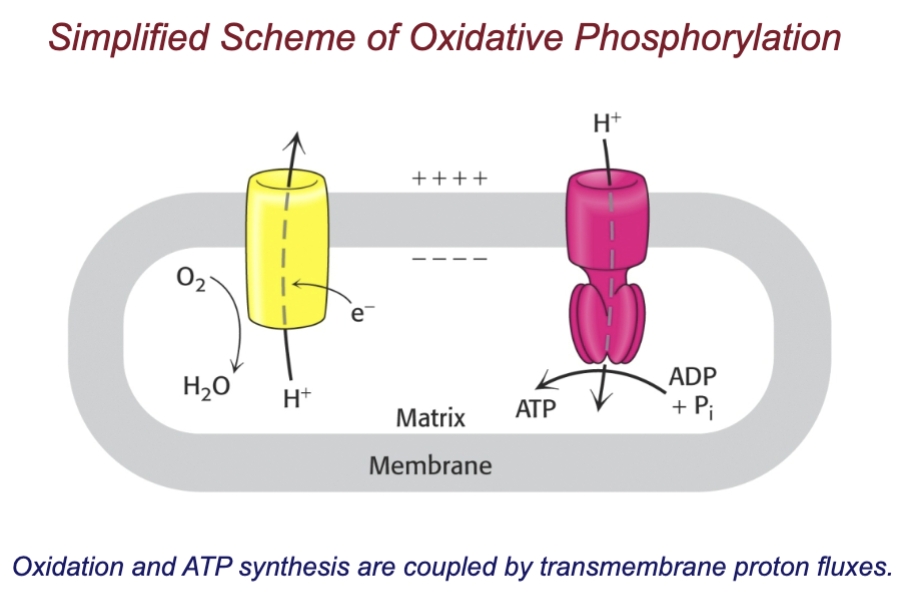
What is the chemiosmotic hypothesis?
ATP is synthesized in mitochondria and chloroplasts by using a proton gradient across a membrane
proton pumping (using electrons) across inner membrane (from matrix to intermembrane space) to create proton gradient (high H+ outside, low H+ concentration inside)
pH gradient and membrane potential provides proton-motive force that catalyzes ATP synthesis
Overview of carbohydrate metabolism and the movement of electrons. Contrast substrate level phosphorylation with oxidative phosphorylation
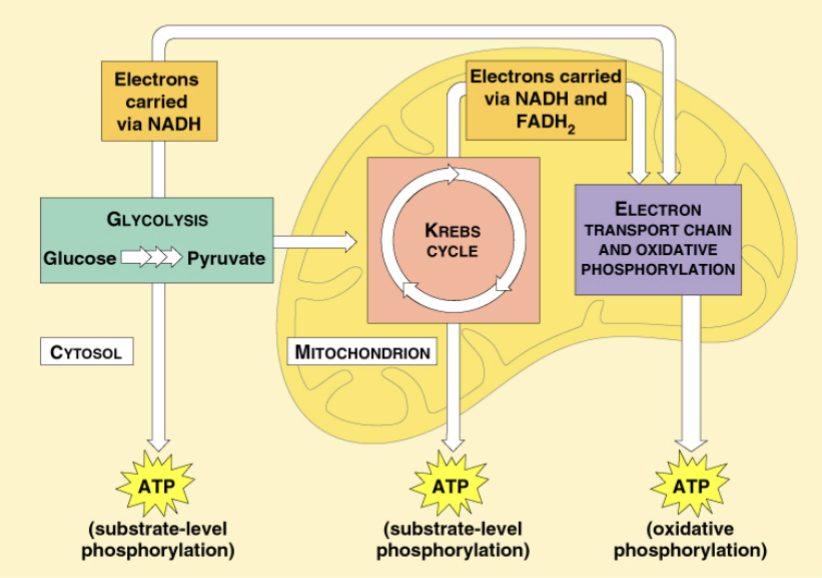
substrate level phosphorylation: forms ATP by transferring phosphate from an intermediate catabolism substrate to ADP
oxidative phosphorylation: forms ATP using energy derived from the redox reactions in ETC
oxidant vs. reductant
oxidant = oxidizing agent = accepts e-
reductant = reducing agent = donates e-
Where in the cell do ETC and oxidative phosphorylation occur?
inner mitochondrial membrane
What does “cellular respiration” include, define it
TCA cycle and oxidative phosphorylation
respiration = ATP-generating process where an inorganic compound (eg. O2) is the ultimate electron acceptor
Chemical equation of the reaction that generates the energy for creating a proton gradient in ETC
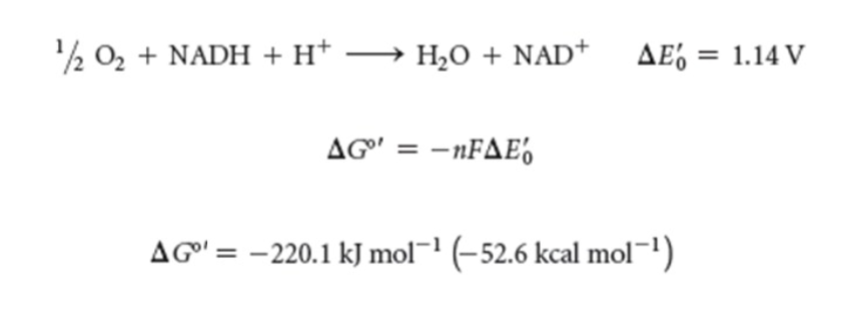
high energy electrons transferred to oxygen —> H2O and energy
the electron transfer potential of an electron is measured as ______. Describe how this value is calculated and understood.
reduction potential E0’ or redox potential
negative E = strong reducing agent, readily donates
positive E = strong oxidizing agent, readily accepts
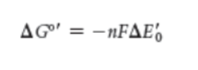
where n = number of electrons transferred and F is faraday constant
What is the condition for standard oxidation reduction potential?
pH 7 25oC
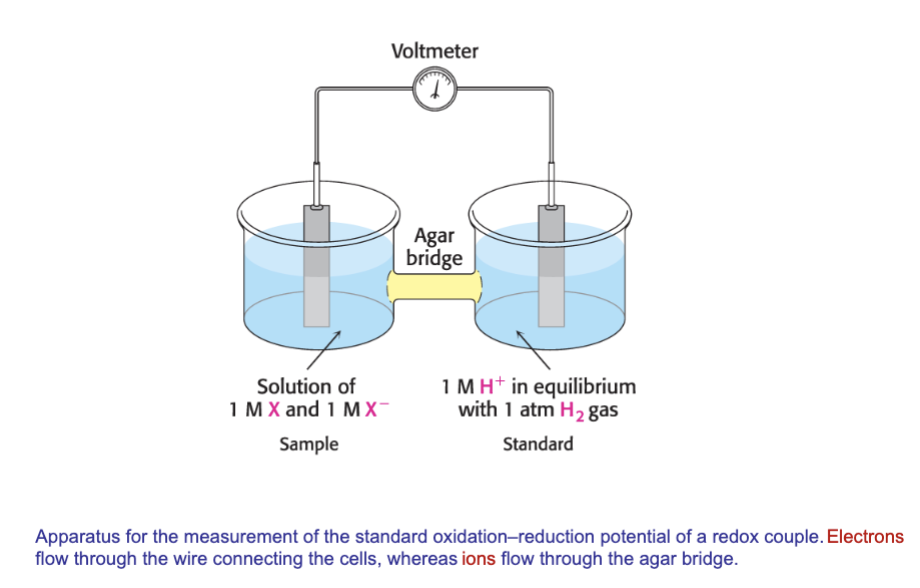
What apparatus can be used to measure redox potential? how does it work?
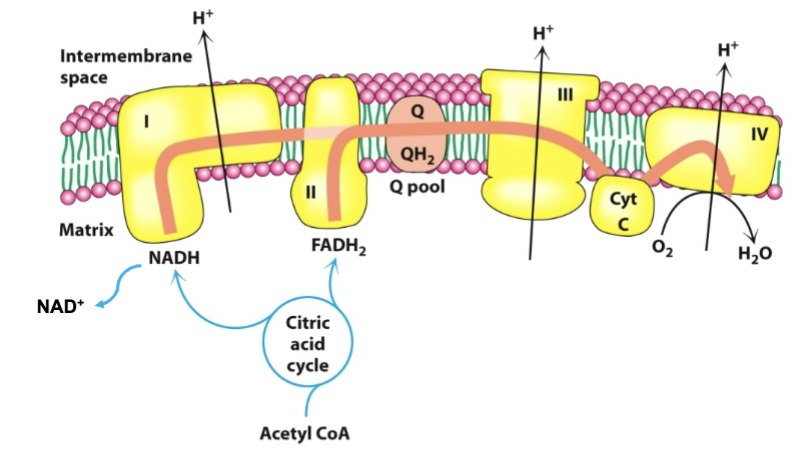
Describe the organization of parts in the ETC (protein complexes, enzymes, electron movement)
four protein complexes in the inner mitochondrial membrane
lipid soluble coenzyme (UQ,CoQ) and water soluble protein (cyt c) shuttle between the four protein complexes
high energy electrons (in the form of NADH and FADH2 from TCA) generally fall in energy from complexes I and II to IV
O2 reduction —> H2O in IV
Describe the change in free energy (relative to O2) across the four proteins in ETC
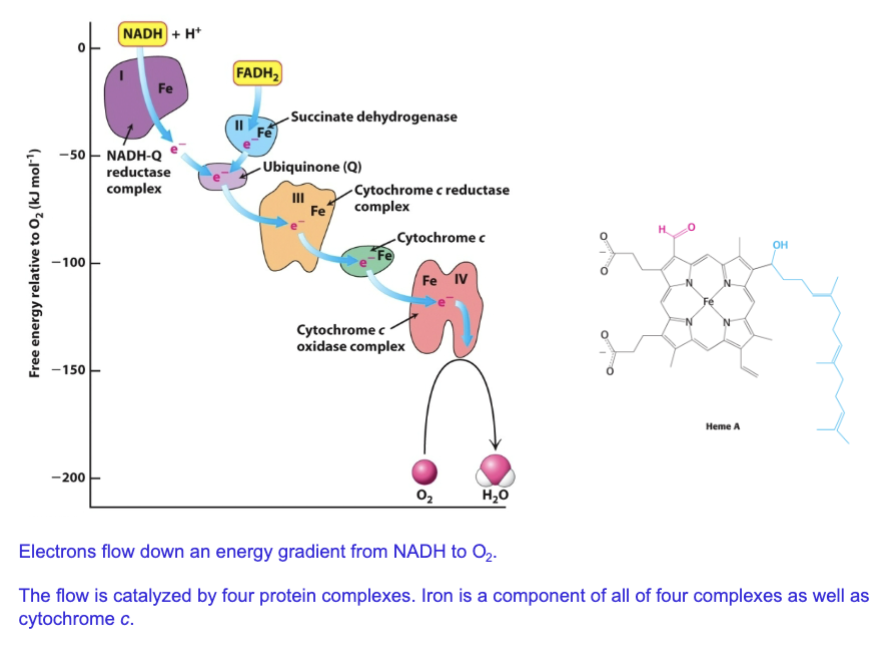
What metal is a component of all four ETC protein complexes (and in cytochrome C)
Iron
Describe the electron affinity Eo’ of ETC components (the four proteins) moving down the chain
electron affinity increases as you go down the chain of complexes
Describe the exact electron + proton passage over the four complexes
electrons reach Q (ubiquinone/Coenzyme Q) through complexes I and II; when going through I, 4H+ is pumped out
QH2 serves as a mobile carrier of electrons and protons, passing e to complex III; 4H+ are pumped out
complex III passes to cytochrome c
complex IV transports e from cytochrome c to O2 —> H2O; 2H+ pumped out
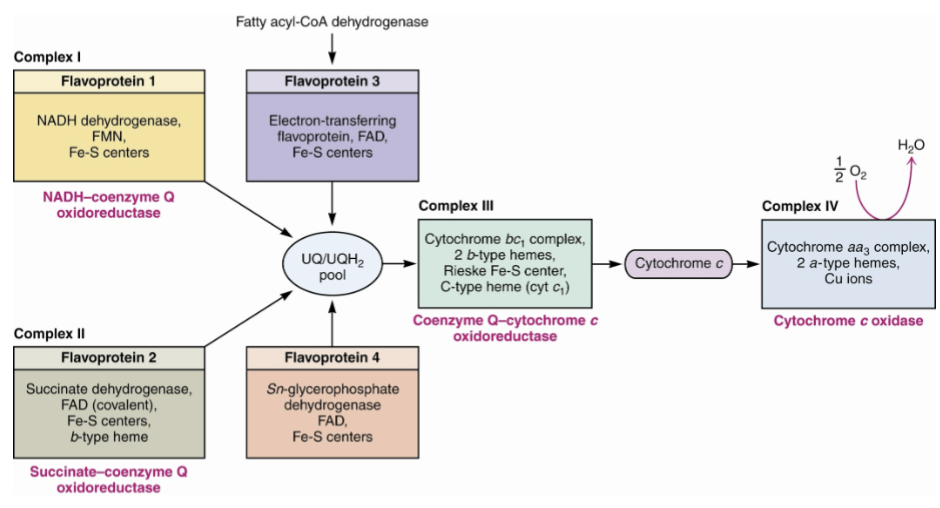
Name complex 1 of ETC, what does it do?
NADH: ubiquinone oxidoreductase
catalyzes 2 simultaneous coupled reactions
H- (hydride) transfer from NADH + H (from matrix) to CoQ (exergonic)
transfer 4 H+ from matrix to intermembrane (endergonic)
—> a proton pump driven by the energy of electron transfer
ubiquinone vs. ubiquinol
different oxidation states of coenzyme Q, the mobile electron carrier
ubiquinol (QH2) is the reduced version (after taking electro from complex I) of ubiquinone (Q)
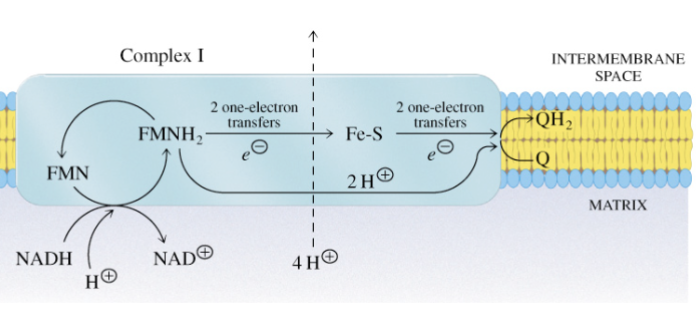
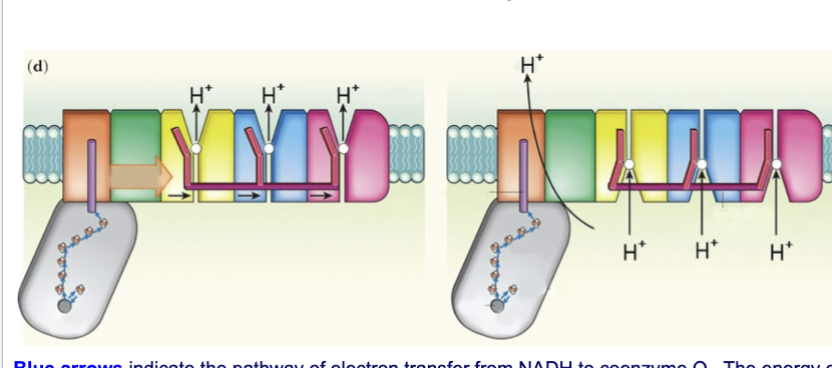
Describe the conformational changes in complex I for transporting protons from the matrix to the cytosol
energy of electron transfer allows proton to cross complex I near the interface between its hydrophilic and hydrophobic domains
—> long helical rod (magenta) reorients subunits to release 3H+ into intermembrane space
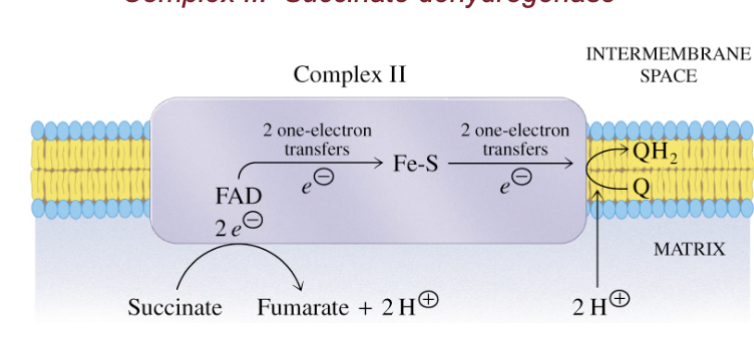
Name complex II and describes its structure and what it does
succinate dehydrogenase
2 prosthetic groups and 4 diff proteins
electrons pass from FAD to an Fe-S center and then to ubiquinone
NO PROTON PUMPING
removes 2 H from succinate to give fumarate + 2H+
reduces (gives electrons) Coenzyme Q
Which complex (I or II) is the entry point for cofactor FADH2 ?
complex II
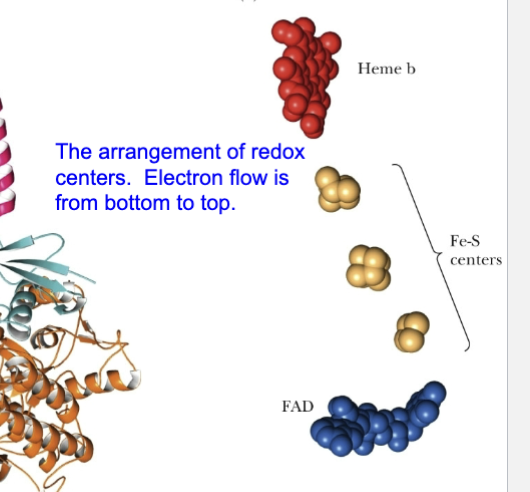
Describe the arrangement of redox centers in complex II
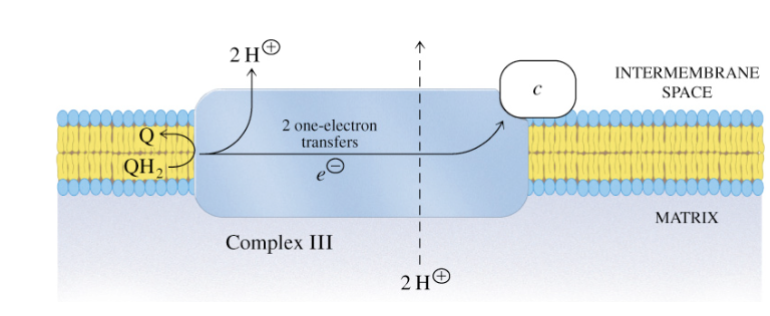
Name complex III and describe its function and structure
Ubiquinone: cytochrom c oxidoreductase
couples the transfer of electrons from ubiquinol to cytochrome c
transfers 4 H+ total into intermembrane
Describe cytochrome c, its structure, and how it transports electrons
soluble protein in intermembrane space
single heme at center of structure, linked to the rest of the protein via 2 sulfur atoms
3rd sulfur from a methionine coordinates the iron
transports electron from complex III to the copper center of complex III
the four ETC complexes may function as supercomplexes, what does this mean?
Supercomplexes are larger, stable associations of two or more protein complexes that are involved in the mitochondrial electron transport chain. These complexes, such as Complexes I, III, IV, and V, are the building blocks of the electron transport chain, which is responsible for generating ATP through oxidative phosphorylation. Supercomplexes are thought to play a role in optimizing the electron transport chain's efficiency and reducing the production of reactive oxygen species
what is the energy stored in the proton gradient called? Describe the two different components of this energy
proton motive force (pmf)
chemical potential energy: difference in concentration of a chemical species in two regions separated by the membrane
electrical potential energy: results from separation of charge when protein moves across the membrane