AUBIO 230: Topic 7 (Protein Sorting & Transport)
1/115
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
116 Terms
difference between eukaryotic cells prokaryotic cells
Eukaryotic cells have membrane bound organelles
intracellular compartmentalization
-Eukaryotic cells use membrane-enclosed compartments to segregate incompatible reactions
-Allows efficiency and specialization
incompatible reactions
Ex. some rxns synthesize glucose while some break down glucose, therefore these reactions have to be separated into organelles
Are incompatible reactions separated in prokaryotes?
Yes, using biomolecular condensates/membrane less organelles
membrane-enclosed organelles
-Eukaryotic cells contain a basic set of membrane-enclosed organelles
-Each has a unique set of molecules and performs a specialized function
-endomembrane system
endomembrane system components
ER, Golgi apparatus, lysosomes, peroxisomes, endosomes
(distinct compartments separated from cytosol by at least 1 selectively permeable membrane)
-relate to protein transport
endomembrane system importance to protein transport
-associated with nuclear envelope & PM
-ER lumen linked to Golgi complex endosomes & lysosomes by transport vesicles that move material b/w them and to/from PM
know function of every organelle in cell
Topic 1
cytosol
contains many metabolic pathways
nucleus
main genome
endoplasmic reticulum
synthesis of most lipids and proteins for distribution to many organelles & to PM
golgi apparatus
modification, sortin, and packaging of proteins & lipids for either secretion or delivery to another organelle
lysosomes
intraclleular degradation
endosomes
sorting of endocytosed material
mitochondria
ATP synthesis by oxidative phosphorylation
chloroplasts
ATP synthesis and C fixation by photosynthesis
peroxisomes
oxidative breakdown of toxic molecules
How did the endomembrane system evolve?
via plasma membrane invagination (ancient archaeal PM formed protrusions that were separated and pushed inwards resulting in membrane-encolced organelels.
How did the mitochondria and chloroplasts evolve?
from endosymbiosis (engulfment of bacteria) -mitochondria (from aerobic bacteria)
-chloroplasts (from photosynthetic bacteria)
protein transport
proteins transported into organelles
3 mechanisms of protein transport
1. transport through nuclear pore
2. transport across membranes
3. transport by vesicles
In which mechanisms do the proteins remain folded (functional structure) and unfolded
remain folded: transport by nuclear pores & vesciles
unfolded: transport across membrane
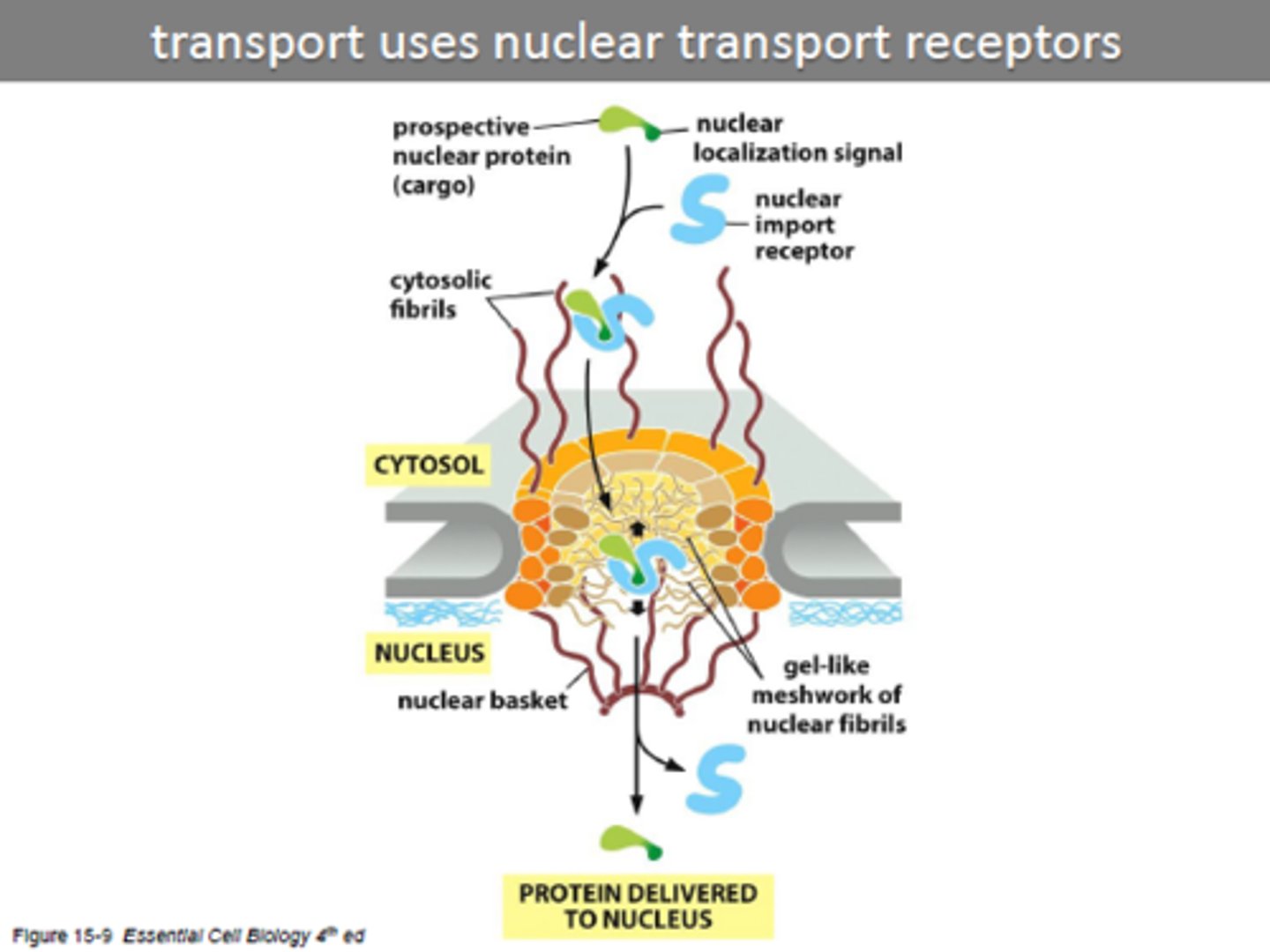
transport through nuclear pore
-occurs via nuclear pores on nuclear membrane allowing exchange of material between cytosol and nucleus
transport across membranes
-protein is unfolded by protein translocators
-guide it across the hydrophobic interior of the membrane
-For endomembrane system organelles, proteins transport via this mechanism to get into those organelles.
transport by vesicles
- proteins & lipids synthesized by ER are passed to golgi to be put into vesicles which fuse to other organelels or to PM
signal sequence structure
short amino acid sequences that direct proteins to their destinations
signal sequence function
-direct proteins to the correct compartment/destination
-can be removed or retained once the protein reaches its destination
Describe how signal sequences work in an example
-Proteins destined for the ER possess an N-terminal signal sequence that directs them to that organelle, whereas those destined to remain in the cytosol lack signal sequence.
-
What determines where a protein goes and where it comes from?
signal sequences
What do proteins enter the nucleus through?
nuclear pores
nuclear pore complex
forms a gate through which selected macromolecules and larger complexes enter or exit the nucleus as protein fibrils protrude from both sides (like a mesh screen)
nuclear localization signal
carried by proteins entering the nucleus from the cytosol
nuclear import receptors
Deliver proteins from the cytosol to the nucleus via the nuclear pores (protein has to bind receptor to be delived to nucleus, cannot enter by itself)
nuclear export receptors
deliver proteins and RNA to the cytosol from the nucleus
-recognize nuclear export signals on such molecules
What drives nuclear transport (materials in and out of nuclease)
powered by GTP hydrolysis
GTP --> GDP Pi
Why do we need to move proteins into mitochondria and chloroplasts?
Although endosymbiotic theory says they both evolved from prokaryotic cells and contain their own ribosomes and dna, most of their proteins come from the cytosol so they need to move into them
What must be crossed for a protein to enter a mitochondria or chloroplast?
inner and outer membrane
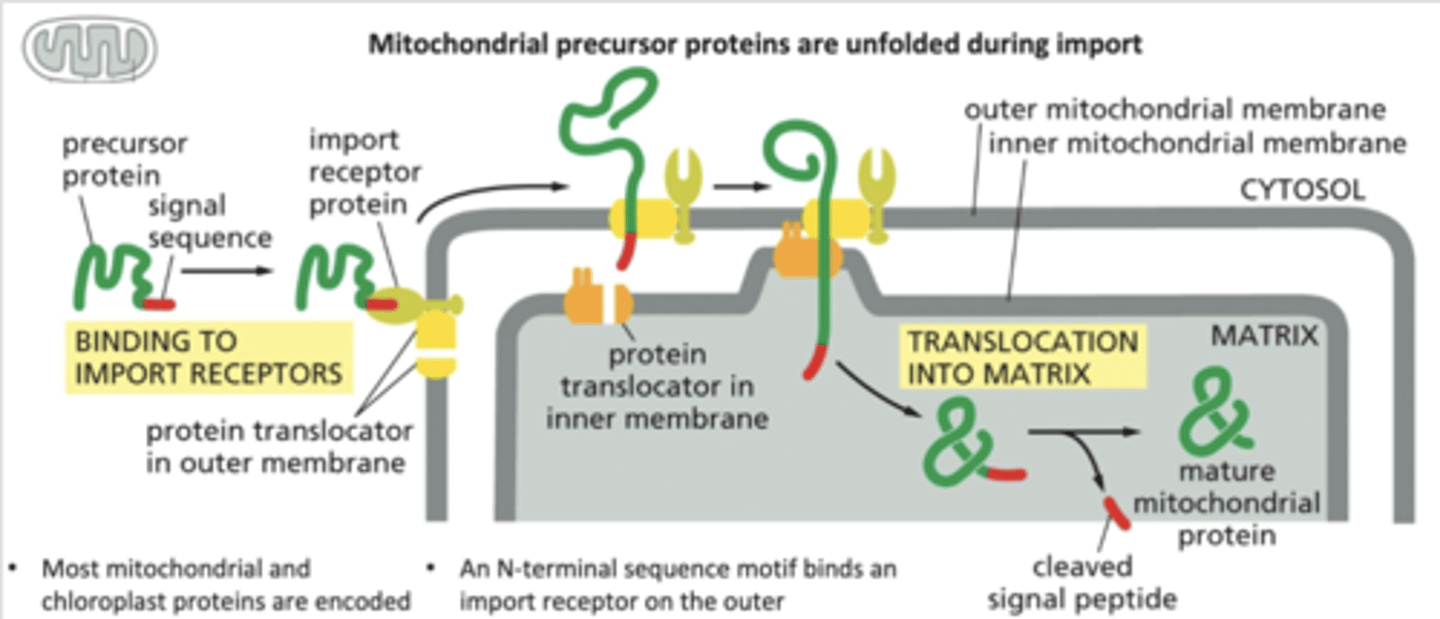
Explain how proteins are transported into mitochondria and chloroplasts
-mitochondrial signal sequence on a mitochondrial precursor protein is recognized by a receptor in the outer mitochondrial membrane & binds.
-receptor associated with protein translocator transports the signal sequence across the outer mitochondrial membrane to the intermembrane space.
-complex of receptor, precursor protein, and translocator diffuse laterally in the outer membrane until the signal sequence is recognized by a second translocator in the inner membrane.
-the 2 translocators transport the protein across both the outer and inner membranes, unfolding the protein in the process
-The signal sequence is finally cleaved off by a signal peptidase in the mitochondrial matrix.
-chaperone proteins help fold protein back
perocisomal transport
-Proteins are delivered from the cytosol via receptor proteins and passed to translocators that to move protein into peroxisome
-Protein is not unfolded moved across as it is
-Some proteins are delivered from the ER via budded vesicles and fuse to the peroxisome
Why is peroxisomal transport different than protein transport
Generally, when proteins move across organelle membranes, they are unfolded. but not in peroxisomal transport
PMP
perisomal membrane proteins (every organelle with membrane has to be renewing the membrane proteins in them)
Transport into the ER
-has 2 pathways
-needs to get its own proteins from cytosol and also export proteins
-involves ER signal sequence, signal-recognition particle (SRP), protein translocator, and SRP receptor
2 pathways/proteins found in the ER
soluble proteins vs. transmembrane (embedded) proteins
ER signal sequence
short amino acid sequence that directs a protein to the endoplasmic reticulum (every protein coming & leaving ER must have)
signal-recognition particle (SRP
binds (by recognizing) to the ER signal sequence and ribosome, pausing translation
protein translocator
membrane channel in the ER through which the polypeptide chain is threaded (controls movement across membrane)
SRP receptor
membrane protein on the ER that binds the SRP, bringing the ribosome to the translocator
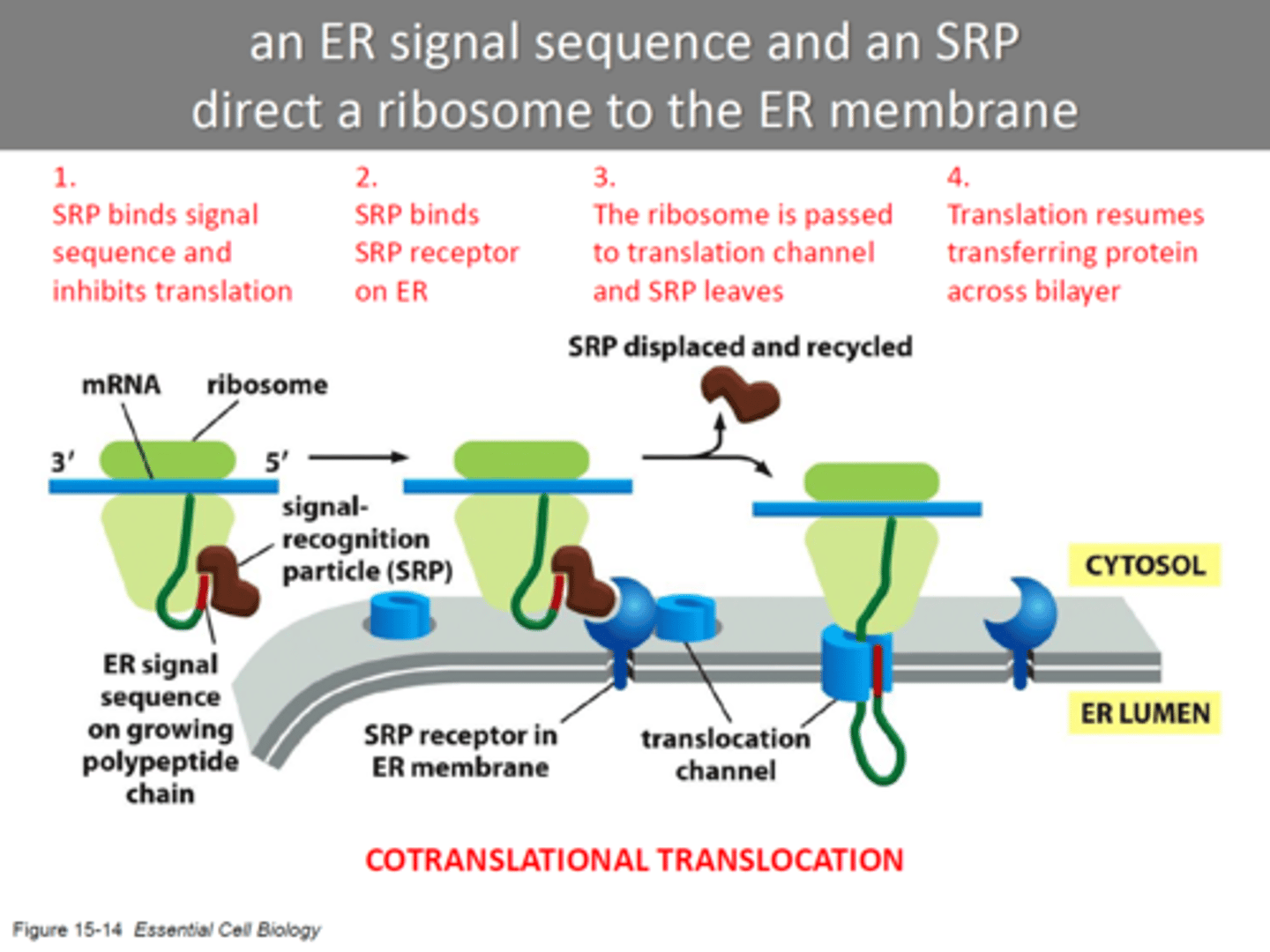
How does the ER signal sequence and SRP (signal recognition receptor) direct a ribosome to the ER membrane?
-ribosome doing translation take protein to ER
-SRP binds to ER signal sequence and the ribosome, slowing protein synthesis by the ribosome.
-SRP-ribosome complex then binds to an SRP receptor in the ER membrane which moves it to translocator.
-SRP is released and reused, and the ribosome passes from the SRP receptor to translocator in the ER membrane and Protein synthesis resumes.
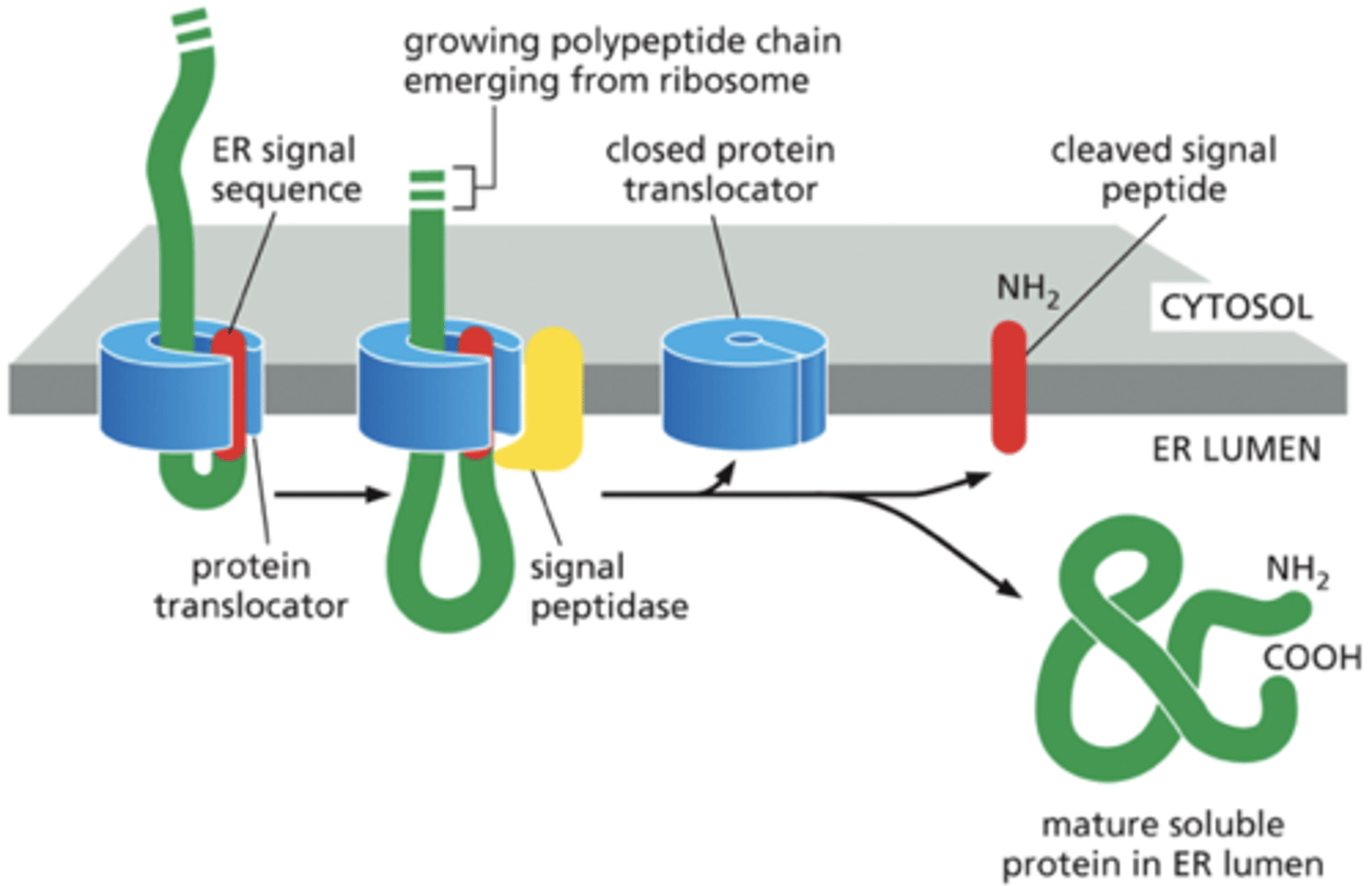
How does a soluble protein cross the ER membrane & enter the lumen
-polypeptide synthesis continues
-protein translocator binds the signal sequence & threads the rest of the polypeptide across the lipid bilayer as a loop.
-signal peptidase cleaves signal sequence to free up polypeptide.
-signal sequence is ejected into bilayer and degraded.
-translocator closes & delivers mature protein into ER lumen
-soluble proteins recite in ER lumen
How is a transmembrane protein retained in the lipid bilayer
-N-terminal ER signal sequence initiates transfer as in soluble protein. 15-14.
-the protein contains a stop-transfer sequence (2nd hydrophobic sequence)
-When this sequence enters protein translocator, the growing polypeptide chain is discharged into the lipid bilayer (halting synthesis)
-N-terminal signal sequence is cleaved off into the lumen and C terminal stays on outside leaving transmembrane protein anchored in the membrane.
-(Protein synthesis on the cytosolic side then continues to completion)
Why do transmembrane proteins stay on the membrane instead of being delivered into the lumen?
stop-transfer sequence stays in the lumen with the ends outside because of lipid bilayer. stop-transfer sequence is hydrophobic, so will interact with hydrophobic tails of bilayer (membrane).
What allows transport between organelles
vesicle budding and fusion
vesicular transport
movement of materials into and out of the cell
-ER --> Golgi
-Golgi --> other organelles, PM
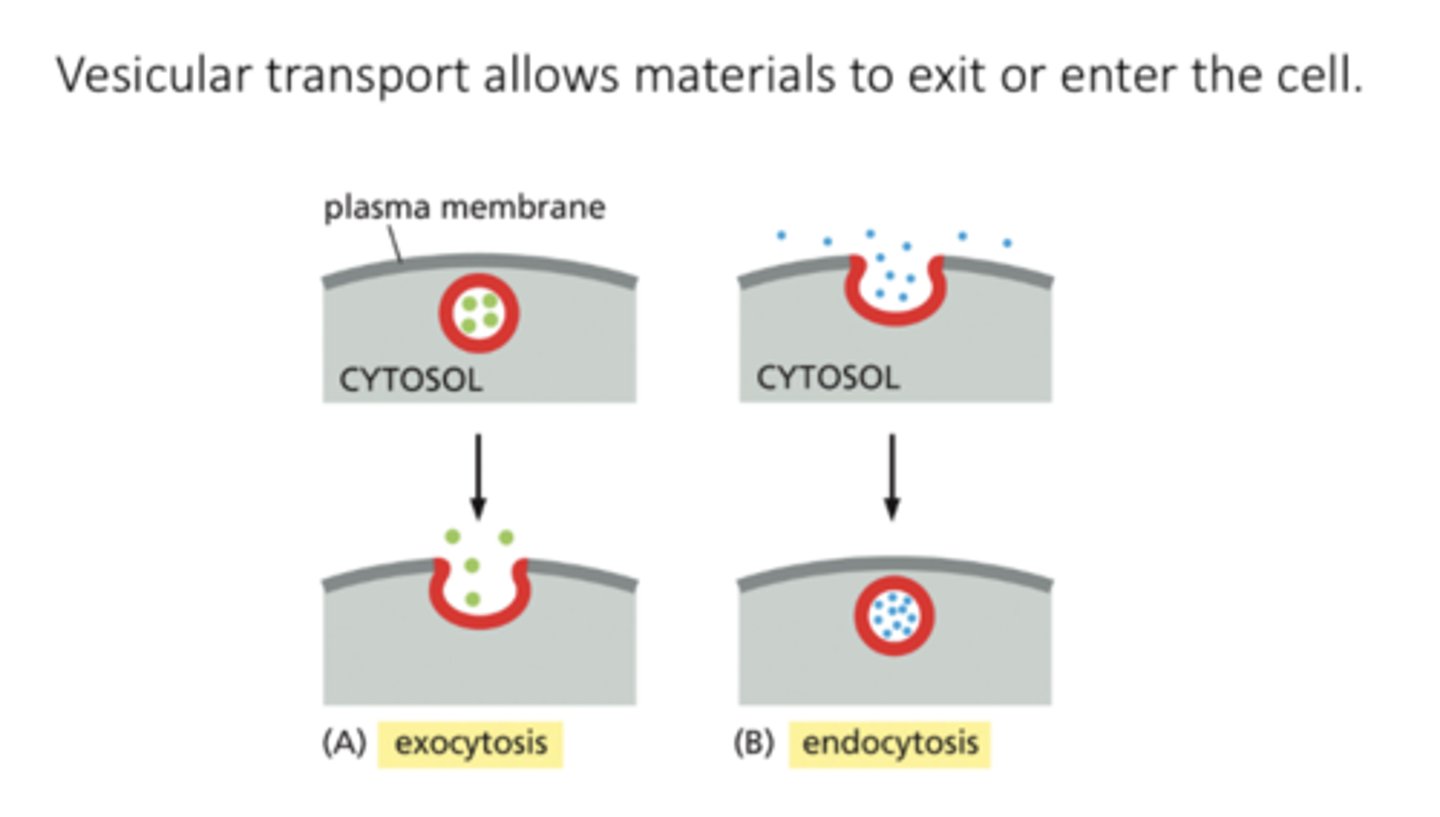
exocytosis
a vesicle fuses with the plasma membrane, releasing its content to the cell's surroundings
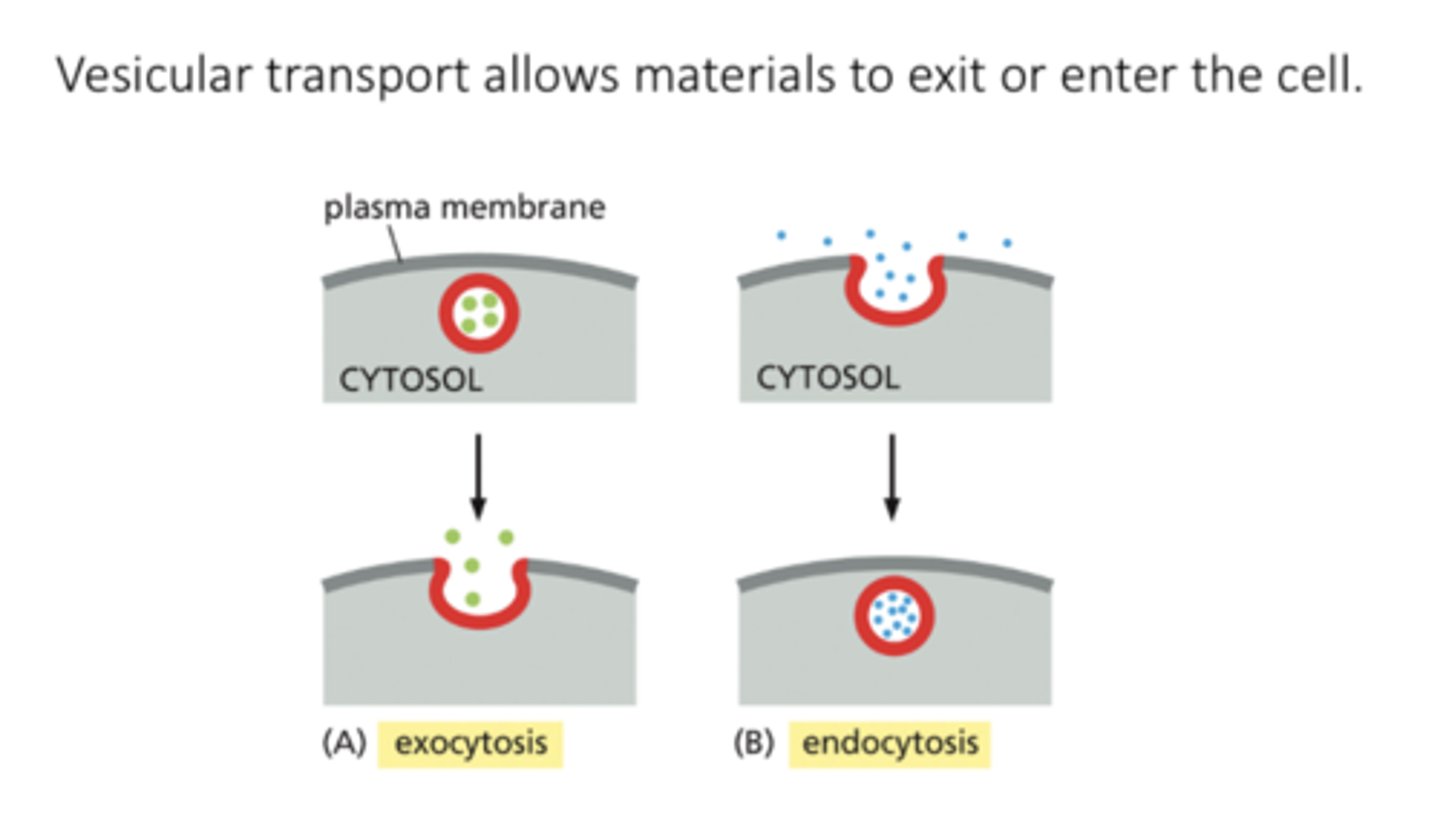
endocytosis
extracellular materials are captured by vesicles that bud inward from the plasma membrane and are carried into the cell
Role of transport vesicles
Transport vesicles carry soluble proteins and membrane between compartments
Importance of transport vesicles
For all endomembrane system organelles, they need to renew their membrane proteins
Process of transport vesicle bud from one membrane fusing with another to carry membrane components between
-membrane of each compartment/vesicle maintains orientation with cytosolic side faces cytsol.
-Endocytosis: vesicles derived from PM delivered to early endosomes and to lysosomes via late endosomes
-exocytosis: protein molecules from ER through GA to PM or to lysosomes.
endosome
organelles that take substance ans pass them to lysosme
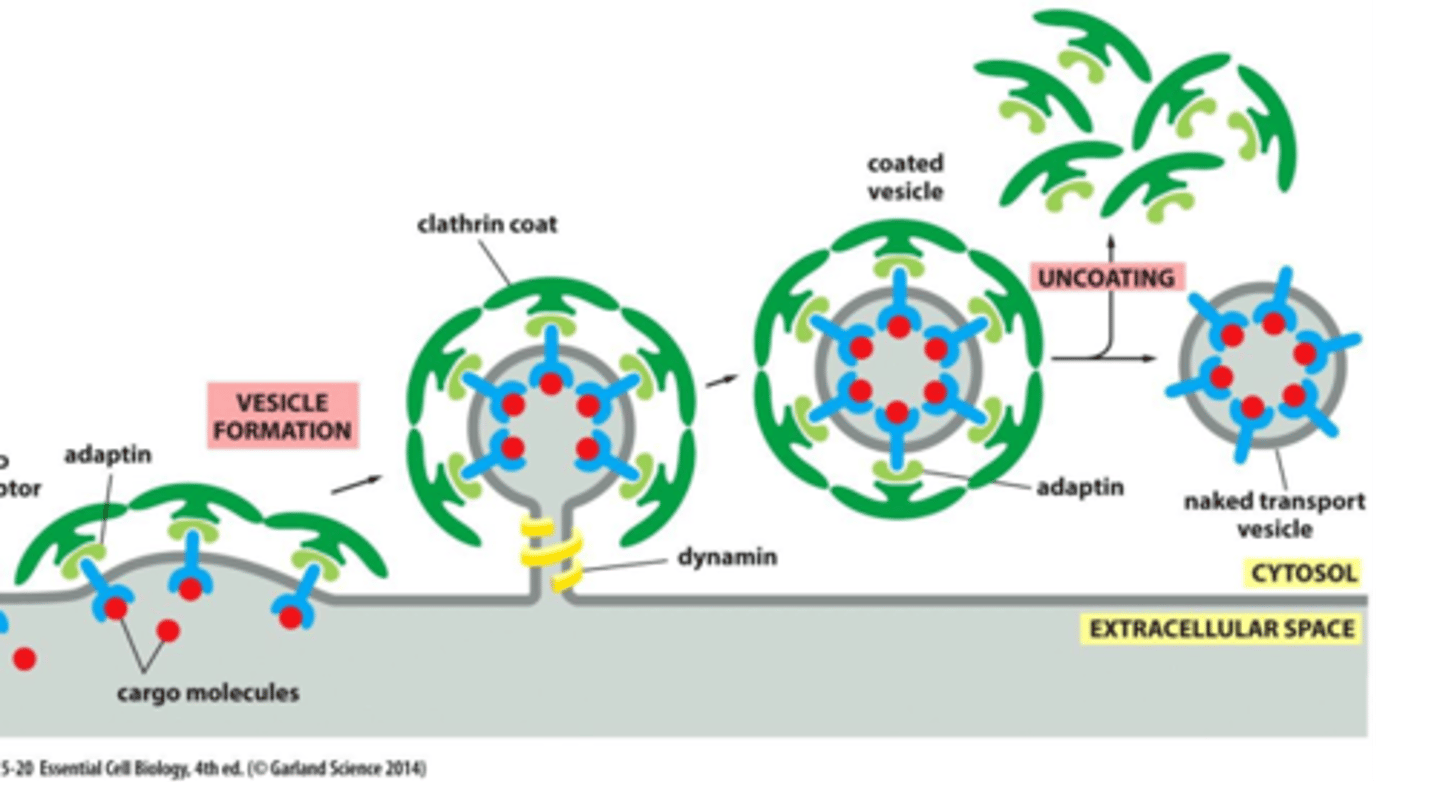
What drives vesicle budding
the assembly of a protein coat
coated vesicles (functions)
shuttle cargo between compartments
Function(s) of protein coat
-shapes the membrane into a bud
-captures molecules for onward transport
Clathrin-coated and COP-coated
Clathrin-coated
-Golgi apparatus -> outward (secretory pathway)
-Plasma membrane -> inward (endocytic pathway)
-Dynamin - pinches off vesicle
-Adaptin - selects cargo molecules
clathrin function
Protects vesicle, gives it a shape, forms basketlike cages around vesicles.
COP-coated
-ER -> Golgi
-one part of Golgi apparatus to another
-E.g., COPI, COPII
How does budding of coating take place?
-Cargo receptor binds to molecules & are captured by adaptins, which also bind to clathrin molecules to the cytosolic surface of the budding vesicle
-Dynamin proteins assemble around the neck of budding vesicles; & hydrolyze their bound GTP pinching off the vesicle.
-After budding complete, coat proteins are removed & reused, and the naked vesicle can fuse with its target membrane/released in cytosol.
Why are different types of coats important?
ave unique adaptins, each responsible for different types of vesicles moving between different compartments of the cell
What does vesicle docking depend on
tethers and SNAREs
Generally when vesicles are moved what happens?
vesicle formation, then ufsion
docking
when vesicle docks on membrane/moves close to it
fusing
when vesicle releases contents inside
What do vesicles display for their identity?
molecular markers on their surface
3 proteins that help direct transport vesicles to their membrane
Rab proteins, tethering proteins, and SNAREs
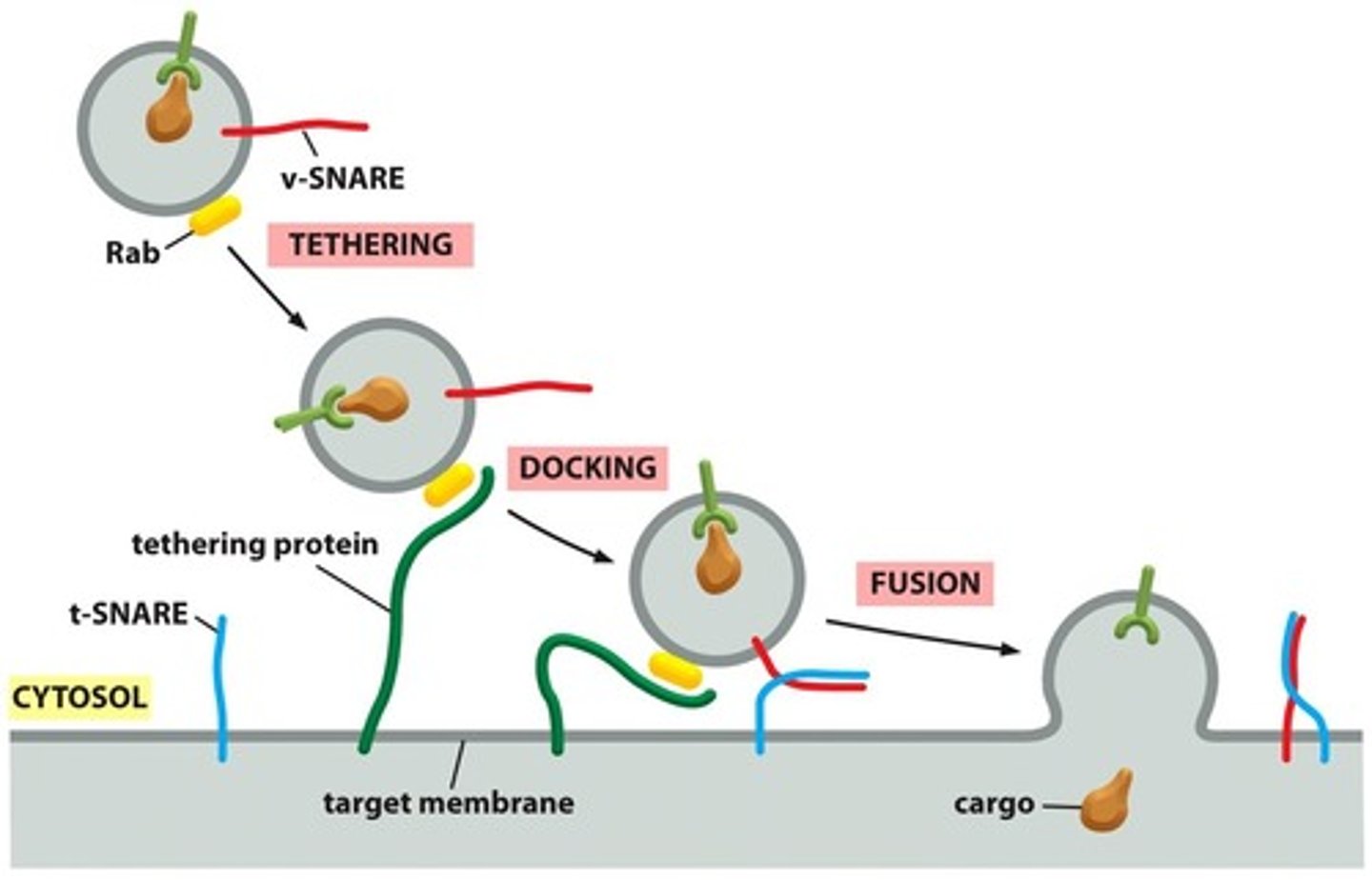
Rab proteins
Organelle-specific markers
-to make sure going to target organelle
tethering proteins
-present on the target molecule (bind to Rab protein to dock on membrane)
Differences between Rab, tethering, and SNARE proteins *
provide the initial recognition between a vesicle and its target membrane, complementary SNARE proteins ensure that transport vesicles dock at their appropriate target membranes.
SNAREs
-help in vesicle docking and membrane fusion
t-snare and v-snare
complimentary and interact to bring vesicle down (docking) to iniate fusion
t-snare
-target membrane snare protein
v-snare
-vesicle
fusion
membranes blend into each other, then contents are released
What catalyzes the fusion of the vesicle and target membranes after vesicle docking
SNARE proteins
Explain the fusion of vesicle to target membrane
-docks by complimentary snares interacting and pulling it down
- force of the SNAREs winding together squeezes out water molecules trapped between the 2 membranes, allowing lipids to flow together to form a continuous bilayer. (membranes blend together)
-contents released into target
-SNAREs are pried apart so that they can be used again
Secretory pathways
path for anything going to the ER to the golgi to the PM/ lysosome
ER modification of protein for secretion
-disulfide bond formation to stabilize protein & protect from degradation
-glycosylation: covalent attachment of oligosaccharide chains (sugar chain) for structural proteins
What occurs when proteins are manufactured?
-some go inside cell
-some go outside cell and outside conditions cause proteins to be degraded
Fate of proteins in the ER
-some proteins are folded correctly so they can function normally, but lots are misfolded (cause disease)
Path of successfully folded proteins in ER
packaged into vesicles
-transported to GA for further processing
-(then released in cytosol?)
Pathway of improperly folded proteins
-retained in ER & assided by chaperone proteins to attempt refolding
-if successful: transported to golgi
-if unsuccessful: exported to cytosol & degraded by proteosome
Unfolded protein response (UTR)
-triggered by accumulation of misfolded proteins in ER
1. ER expands
2. slows down protein synthesis
3. increases production of chaperones
goal of unfolded protein response (UPR)
restore ER function and reduce stress
How does ER decide to use a component of the UPR, such as producing more chaperones?
-misfolded proteins recognized by transmembrane sensor proteins in ER membrane each of which activates a different component of the UFR
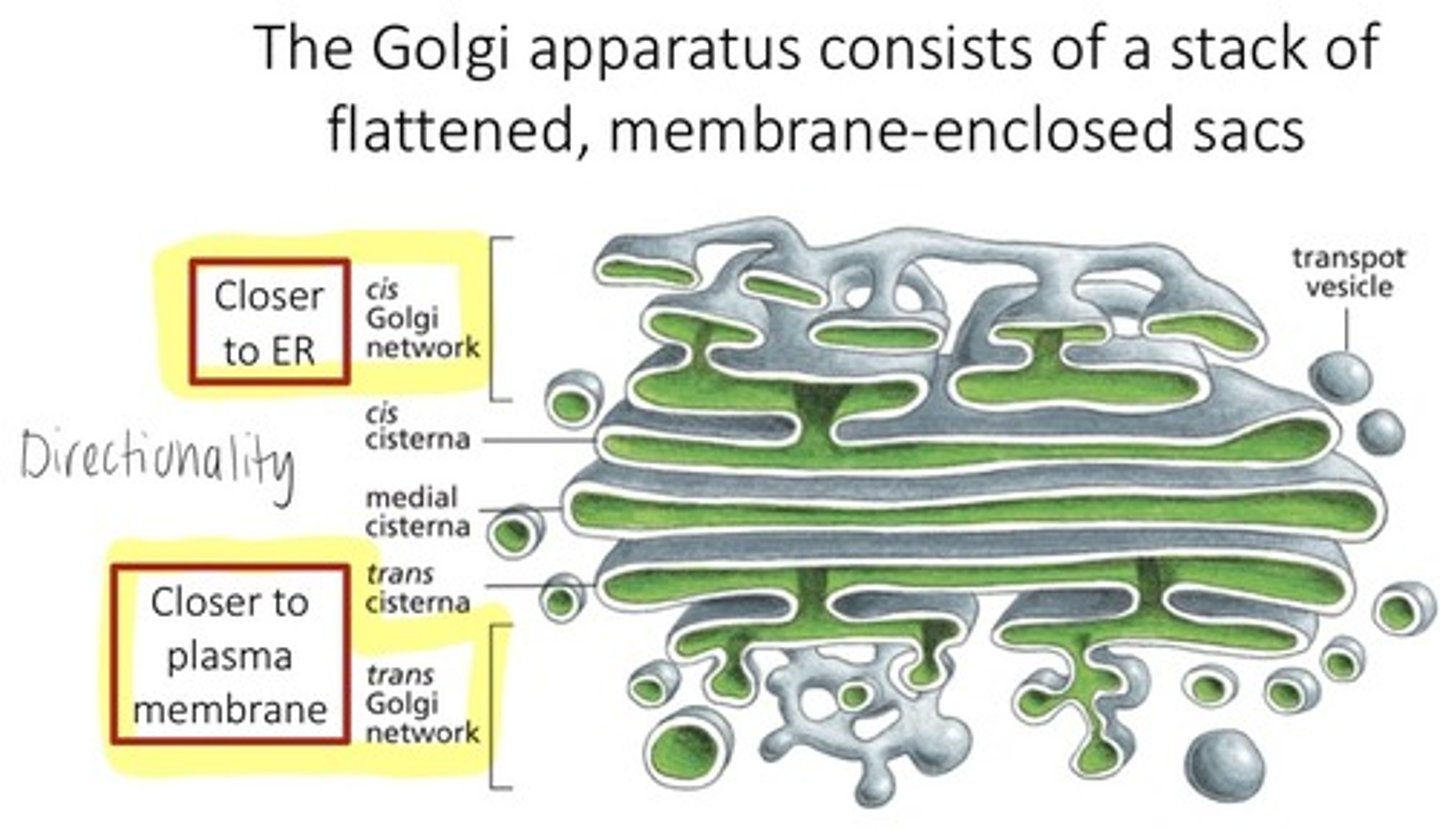
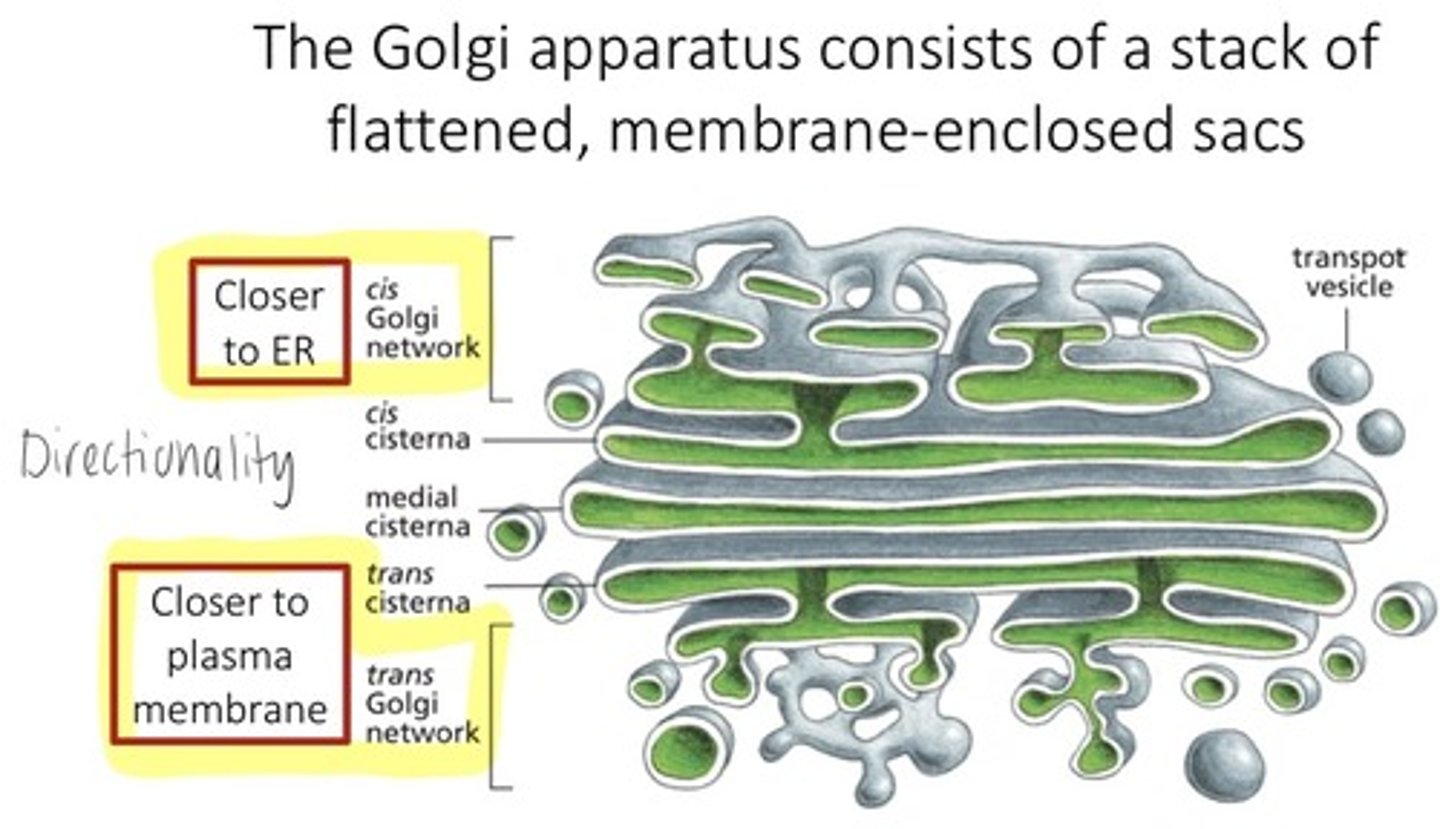
where are proteins further modified and sorted
golgi apparatus
Golgi apparatus role
protein/lipid modification, sorting, packaging
sorting (by GA)
directs each vesicle to where its supposed to go
2 sides of golgi apparatus
ER side: cis
Trans: faces PM
Vesicles released from golgi apparatus
via exocytosis, vesicles passed to endosome, then to lysosome where it is degraded or it could go to the plasma membrane
What are secretory proteins released from the cell by?
ecocytosis
2 pathways (types) that secretory preotiens are released from the cell via exocytosis
Constitutive and regulated