BIO367 - Concordia University
1/85
Earn XP
Description and Tags
Dr.Andrew Wieczorek - Winter 2025
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
86 Terms
What are the characteristics of RNAPI?
found in nucleolus
transcribes large precursor to major rRNAs
transcribes one gene
What are the characteristics of RNAPII?
found in nucleoplasm
transcribes precursors to mRNAs (hnRNA + snRNAs)
What are the characteristics of RNAPIII?
found in nucleoplasm
transcribes precursors for 5srRNA and tRNAs
When cloning a plasmid using a single cut site, what is the vector treated with before ligation?
Alkaline phosphate
removes 5’ phosphate that ligase needs to self-ligate back onto itself
When using DNA footprinting to identify different promoters, what would one expect to see from a class I promoter and what are the elements called?
one core element and an upsteam element (UPE) about 100-150bp upstream of transcription initiation

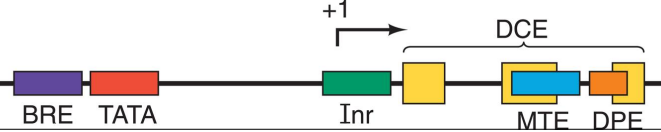
When using DNA footprinting to identify different promoters, what would one expect to see from a class II promoter and what are the elements called?
BRE - binding site for TFIIB (-37 to -32)
TATA - binding site for TBP (-26 to -31)
Inr - binding site for TFIID (-2 to +4)
DCE - binding site for TFIID (+6 to +33)
MPE (within DCE) - downstream promoter element
MTE (within DCE)
When using DNA footprinting to identify different promoters, what would one expect to see from a class III type I promoter and what are the elements called?
5S rRNA
Box A, intermediate element, Box C

When using DNA footprinting to identify different promoters, what would one expect to see from a class III type II promoter and what are the elements called?
tRNA
Box A, Box B

When using DNA footprinting to identify different promoters, what would one expect to see from a class III type III promoter and what are the elements called?
snRNA gene
DSE (distal sequence element), PSE (proximal sequence element), TATA box

What is the order of binding DNA to create the preinitiation complex.
TFIID, TFIIA, TFIIB, RNA Polymerase II, TFIIF, TFIIE, TFIIH
Where do the TBP/TFIID bind DNA?
The minor groove
What is TFIID
TBP (TATA binding protein) + TAFIIs
Why are TAFIIs important?
They bind the Inr
They bind the DPE
They bind to other factors important for transcription
What is the last TF to join the preinitiation complex?
TFIIH
What is the function of TFIIH?
It is a helicase - unwinds DNA at transcription site
TFIIH phosphorylates the C-terminal domain of RNAPII and is stimulated by the presence of TFIIE
What is the function of TFIID
Has TBP, first TF in preinitiation complex
What is the function of TFIIF?
Aids in polymerase recruitment - does not bind have binding site for DNA
Does directionality matter in enhancer sequences?
no
Where does TFIIH phosphorylate RNAPII?
C-Terminal Domain (CTD)
What does TFIIS do?
stimulates stabile transcription elongation
Proof-reading activity of transcript
What does TFIIIC do?
It binds Box A and Box B
Recruits RNAPIII
Why is the TATA box important
gives location of transcription start site + transcription efficiency
Why is the large intron of most class II genes important?
There is an enhancer within this intron that is needed to generate mRNA
Is the TATA box an enhancer?
no, it is a promoter
Where on DNA does the TBP bind?
The minor groove
In what situations are TAFIIs more important for transcription
more important in genes with a Inr or a DPE
less important in genes with just a single TATA box
Is TBP still required for transcription even if there is no TATA box?
yes, it is within TFIID which also has other TFs which need to be assembled and present for transcription to occur
TBP may not bind to DNA, but other TFs are bound to it which may interact with other TFs that are bound to DNA
What is the order of TF binding in type III promoter regions?
TFIIIC (binds Box A and Box B), TFIIIB (contains TBP), Polymerase (TFIIIC falls off during transcription as to not block polymerase)
What are activators?
Gene specific transcription factors
What are the 3 domains of an activator?
DNA binding domains
Transcription Activation Domains
Dimerization Domains (sometimes not needed)
What is the charge of DNA?
negative
What is SP1 and what does SP1 bind to?
SP1 is a C2H2 zinc finger activator for class II promoters
binds TATA-less promoters —> GC boxes
What kind of transcription factor is Gal4?
Activator
What is the role of Galactose in the Gal operon?
Galactose binds Gal80 which relieves repression of Gal4
What does Gal80 do in the Gal operon?
Gal80 is the repressor for Gal4
The different expression pattern of the yeast Gal1 is determined by its gene’s enhancer/UAS region
yes
What was does genetic imprinting say?
the genes expressed by offspring expressed by the methylated gene passed down by paternal mouse
By what mechanism does activation occur?
Enhancer functions to bind activators, activators use looping mechanism to recruit general transcription factors and RNAP promoters
What does ‘squelching’ mean?
One activator inhibits another activator from working by competing for the same binding site
Do mediators bind DNA?
no
What is heterochromatin?
dense, silenced DNA
packed tight
What is euchromatin?
loose DNA almost always under transcription
What subunits are the core histone made of?
H2A, H2B, H3, H4
In histones, what is H1?
The linker protein
What is the charge of the histone tails?
Positive - binds to DNA because DNA is negatively charged
In histone modification, what amino acid is modified during acetylation?
Lysine
What is the relationship between activators and histones?
Histone and activators compete
Activators act as both activators and antirepressors
What does histone acetylation do to the interaction between histones and DNA?
weakens the interactions between DNA and the histone
What does histone deacetylation do to the interaction between histones and DNA?
strengthens the interactions between DNA and the histone
What protein complexes are required for chromatin modifications for gene activation?
Histone Acetyltransferases (HATS)
Chromatin Remodeling Factors (SWI/SNF)
Where does Rap1p bind on DNA?
Rap1p binds to the telomeres
What class of genes does the spliceosome act on?
Class II genes
The first 2 bases of an intron are
GU
The last 2 bases of an intron are
AG
Can the branch point ‘A’ in an intron be any A within the intron?
No, requires consensus sequence
What are spliceosomes made of?
Snurps
U1, U2, U4, U5, U6
In the spliceosome, what does U1 recognize and bind?
5’ splice site of intron
In the spliceosome, what does U2 recognize and bind?
The intron branching point of intron
What is U1 nudged off by in the process of the spliceosome?
U6 (associated with U5)
In the spliceosome, where does U5 recognize and bind?
Complex of U1 and U2 (not recruited but comes along after these have been assembled)
Is U1 in the spliceosome sequence specific?
yes, mutation causes lack of splicing
countermutation to create complementarity yields splicing
Is U6 in the spliceosome sequence specific?
yes, mutation causes lack of splicing
needs UGU on pre-mRNA to pair with U6 ACA
Is U5 in the spliceosome sequence specific?
No
Is U2 in the spliceosome sequence specific?
yes
What does the mechanism of group I self-splicing introns look like?
Sequence of intron made such that it adopts a tertiary structure and a nucleophilic attack occurs which cleaves the intron without the need for spliceosome
What does the mechanism of group II self-splicing introns look like?
-OH group on branch point adenine performs same double nucleophilic attack to splice itself without need for spliceosome
creates lariat structure
What is the difference between cap 0, cap 1, and cap 2
Cap 0 has no methyl groups, cap 1 has 1 methyl group on the first nucleotide, and cap 2 has a methyl group on both the 1st and 2nd nucleotide
What is the order of capping for cap 2
RNA triphosphate clips gamma-phosphate (leaves just aB phosphates)
Guanylyl transferase attaches GMP (reestablishes 3 phosphates on Guanine aBa)
methyl transferase adds Met to guanine cap
methyl transferase adds Met to guanine cap
What processes are 5’ caps required for?
Intron removal
mRNA transport out of nucleus
translation by ribosomes (CAP binding protein)
mRNA stability (protect mRNA from RNAse)
What does the 3’ poly-A tail do?
protects mRNA from RNAse
What happens at the GU/U element at the 3’ end of mRNA after the polyadenylation signal?
GU/U element is bound by CFs (cleavage factors) to cleave site for polyadenylation
What does the polyadenylation signal look like/do?
AAUAAA + GU/U element after
marks site for poly-A tail
What is the rate of polyA degradation determined by?
signal in 3’ UTR
How does the CoTC site contribute to transcription termination?
causes cleavage of the RNA downstream of the poly(A) site. This leaves a free 5′ end that Xrn2 can bind and chew up. As Xrn2 moves along the RNA, it catches up to RNA polymerase II and helps terminate transcription by "torpedoing" it off the DNA
What does DICER do?
enzyme that cleaves dsRNA into siRNA
What is rho dependent termination?
rho binds to polymerase and waits for rho dependent termination site — then rho dissociates transcript from polymerase
What is the DNA characteristic of a rho dependent termination site?
inverted repeat
What is the mechanism rho independent termination
Symmetrical repeat creates a hairpin followed by a string of U’s
transcription terminated after string of U’s
what does β-galactosidase do?
Breaks lactose into galactose and glucose
What is the inducer of the lac operon?
Allolactose
What gene in the lac operon codes for cAMP?
cyaA
In the lac operon, what is the effect of high glucose on cAMP?
When glucose is present, cAMP production is low
Where does the CAP-cAMP complex bind to RNA polymerase?
α-subunit
How does RNA polymerase moving down DNA not create supercoiling?
topoisomerase
nontemplate strand =
coding strand
What would happen if a new sigma factor was incorporated into an RNAP?
the RNAP would recognize a different promoters