09 - Post-transcriptional Regulation
1/70
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
71 Terms
Post-transcriptional regulation
Post-transcriptional control is everything that happens at the level of the RNA.
Includes processing, export, stability vs degradation, modifications, localization, and initiation of translation
what does Post-transcriptional regulation influence
influence how much protein is made from an RNA transcript
Can also influence the protein sequence (Ex. alternative splicing)
Regulation of Alternative Splicing
Splice isoforms can have different activities/functions in the cell
Exonic splicing enhancer elements and exonic splicing suppressor elements are sequences for recruitment of regulatory proteins and spliceosome components
Splicing can be regulated so that particular tissues contain particular splice isoforms
Regulation of Alternative Splicing - positive control
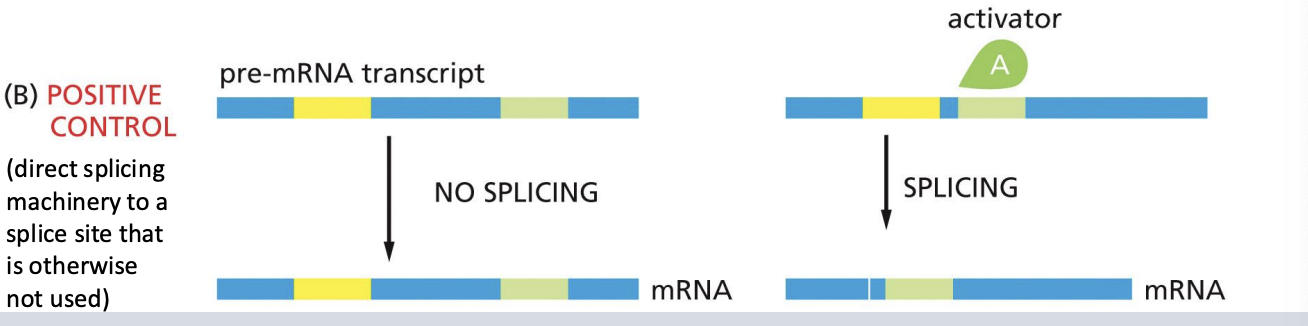
Regulation of Alternative Splicing - negative control
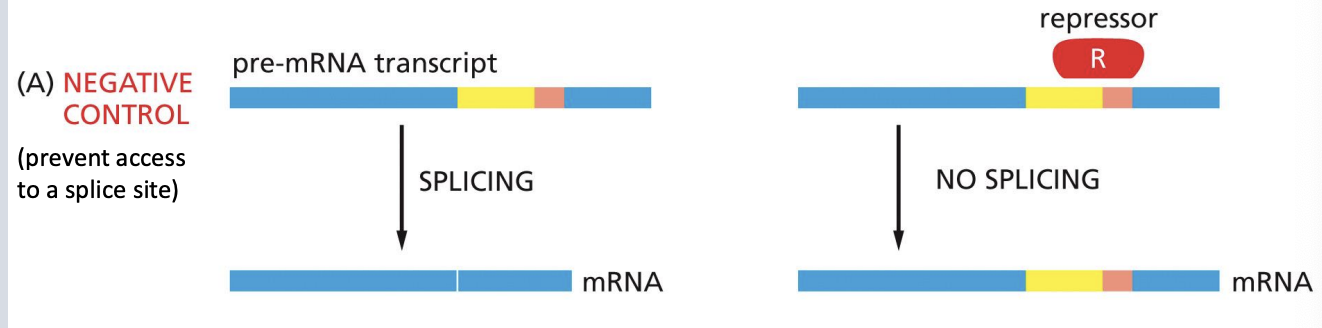
why some alternative splicing could be constitutive
• If splicesome cannot consistently distinguish correct pairings of 5’ and 3’ splice sites, can result in constitutive alternative splicing in all cells
purpose of nonsense-mediated decay
Prevent translation of faulty mRNAs with premature stop codons.
Mechanism of nonsense-mediated decay
Splicing leaves behind exon junction complexes (EJCs). If a stop codon appears upstream of an EJC, NMD machinery degrades the mRNA.
What other mechanisms help ensure protein is produced from correctly processed mRNA?
control over which mRNA are exported from the nucleus
eIF proteins connect the 5’ cap and poly-A tail before initate translatetion
How could covalent modifications and RNA editing impact the structure and function of an RNA molecule?
can create or destroy binding sites for proteins
can cause condons to base-pair with a different tRNA
can influence translation initiation
can change RNA secondary structure
can influence RNA stability
how can covalent modification of mRNA influence the amount and amino acid sequence of a protein
Modifications can:Change mRNA stability. Affect translation efficiency. Influence structure, altering protein-RNA or RNA-RNA interactions.
RNA editing (e.g., A-to-I or C-to-U) can change codons, altering amino acid sequence of the resulting protein.
regulatory mechanisms that impact cleavage and polyadenylation
Many genes contain multiple polyadenylation sites: a weak one upstream and a strong one downstream.
regulated by polyadenylation signal strength and the concentration of processing factors like CstF
example of cleavage and polyadenylation
In a resting B cell, low levels of CstF favor cleavage at the stronger downstream site.
This includes additional exons and results in a longer mRNA with different coding sequences at the end.
The resulting protein has a membrane-anchoring domain, so the antibodies are membrane-bound.
In an activated B cell, high levels of CstF lead to use of the upstream weaker site.
This causes early cleavage and results in a shorter mRNA.
The protein lacks the membrane domain and instead is secreted from the cell.
Which end of the mRNA & protein does Alternate cleavage & polyadenylation alter?
the 3’ and c- terminus
how cleavage and polyadenylation would alter the protein
Alters the C-terminal (3′ end) sequence of the protein.
This changes protein localization and function
Which of the following correctly describe mechanisms that could create slightly different versions of proteins from a single gene?
A) Leaky scanning for Kozak sequences can create versions of the protein that differ at the N-terminus
B) Alternative splicing can create splice isoforms with different amino acid sequences
C) Methylating CpG in promoter sequences can create versions of the protein that differ at the N-terminus
D) Alternative cleavage and polyadenylation can create versions of the protein that differ at the N-terminus
E) RNA editing can create versions of the protein with different amino acid sequences
a, b, e
5’ and 3’ UTRs
Specific sequences in the UTRs determine the interactions of mRNAs with regulators and with translation machinery
Three main types types of control acting through UTRs:
1. mRNA stability (half-life of mRNA determines how long the message is present to be used for translation)
2. mRNA translation (yes/no, when, how often)
3. mRNA localization (where in the cell)
Regulation of mRNA stability - Deadenylation-Dependent Decay
In the cytoplasm, deadenylases gradually shorten the poly-A tail of the mRNA.
Once the tail is too short, the mRNA becomes unstable and is targeted for degradation.
This process is regulated, not random:
Different deadenylation pathways act on specific mRNA sets.
Rescue mechanisms exist to re-extend the poly-A tail, reactivating translation of silenced mRNAs.
Regulation of mRNA stability - Role of the 3′UTR
Sequences in the 3′UTR recruit proteins that control the length of the poly-A tail.
Changing the 3′UTR (e.g., through alternative splicing or mutation) can alter mRNA half-life and protein output.
Regulation of mRNA stability - Endonuclease-Mediated Decay
mRNA degradation can also occur through recruitment of endonucleases to specific internal sequences
These cis-acting sequences are often in the 3’UTR of an mRNA
Blocking or exposing the endonuclease site regulates mRNA degradation
nucleases
(enzymes that breakdown nucleic acids)
the difference is between an endonuclease and an exonuclease?
endonuclease cut within nucleic acid chain
exonuclease remove a single dNMP from the ends of one dna strand
Regulation of translation in prokaryotes
In bacteria, exposing or blocking the Shine-Dalgarno sequence can turn translation on and off
Again, can have trans-acting regulators and cis-acting sequences
The 3D structure of an RNA is often important for allowing or inhibiting protein binding (temp can cahneg it )
Regulation of mRNA localization
mRNAs can be localized to specific regions of the cytoplasm.
Localization is regulated by RNA-binding proteins
These proteins help transport or anchor the mRNA to specific sites within the cell.
Regulation of localization typically occurs through
regulators binding to sequences in the 5’ and 3’ UTRs
How would mRNA localization impact the protein produced from an mRNA?
mrna localization influences where in the cell a protein is syntheiszed (which can affect its function, interactions, and localization)
mechanisms for regulating mRNA localization
1. Diffusion + Anchoring (Trapping)
2. Diffusion + Localized Protection
3. Active Transport
1. Diffusion + Anchoring (Trapping)
mRNA randomly diffuses throughout the cytoplasm.
When it reaches the target area, it is anchored or trapped by localized binding proteins or structures (e.g., cytoskeleton, organelles).
This prevents further movement and concentrates the mRNA where translation is needed.
2. Diffusion + Localized Protection
mRNA diffuses throughout the cytoplasm, but only in specific regions is it protected from degradation.
Elsewhere in the cell, degrading enzymes destroy unprotected mRNAs.
This results in localized accumulation of intact mRNA where protection exists.
3. Active Transport
mRNA is actively transported along the cytoskeleton (e.g., microtubules) using motor proteins.
This is a directional and energy-dependent process.
Ensures precise delivery of mRNA to specific cellular regions, often over long distances in large or polarized cells (e.g., neurons, oocytes).
Controlling cell fate & function by mRNA localization
- Allows localized translation at sites where a protein is in high demand
- Allows unequal distribution into daughter cells during division to create 2 cells with different fates (ex. a stem cell divides to selfrenew and produce a differentiating cell)
why is regulating mRNA localization important for development/function
Both proteins and mRNA are unequally distributed to help set up organism body plans and create daughter cells with different fates
Some mRNA (through their 3’UTR) are even tethered to cell division machinery to ensure they end up in particular cells
Allows asymmetric cell division
Describe mechanisms that could result in different versions of a protein expressed (from the same gene) in different cells
Alternative splicing (changes exon composition).
Alternative promoters (changes 5′ end).
Alternative polyadenylation (changes 3′ end).
RNA editing (alters sequence post-transcriptionally).
Leaky scanning (different start codons).
Define maternal contributions
are proteins and RNA that are stored in the egg until they are needed to orchestrate early development (and determine the fate of particular cells)
describe how a large proportion of maternal contributions could be degraded at the same time and how is it possible
In zebrafish, miR430 is transcribed in early development and targets ~40% of maternal contribution mRNA for degradation!
a sequence in the utr
Why might degrading a large portion of maternal contribution be useful during development?
for transitioning control from maternal to the embryo’s own gene expression
Regulating mRNA with siRNA
miRNA are expressed from the genome, but double - stranded RNA from exogenous sources (siRNA) can also initiate silencing by RNAi
miRNA evolved from siRNA
how does exposing a regulatory sequence impact protein production from an mRNA
Exposing a stabilizing element = increase mRNA stability = more protein production
Exposing a degradation signal or microRNA binding site = faster mRNA decay = reducing protein output
Exposing a translational enhancer = May increase translation= more protein.
how does blocking a regulatory sequence impact protein production from an mRNA
Blocking a degradation signal or miRNA binding site = Protects mRNA from decay = increased protein
Blocking a ribosome binding site or translation start site = Prevents translation initiation = reduced or no protein production
how does deleting a regulatory sequence impact protein production from an mRNA
Deleting a stabilizing or localization signal = result in mislocalization or instability = reduced protein
Deleting an inhibitory element (e.g., repressor binding site) = increase translation = more protein
Deleting polyadenylation signals or splicing enhancers = abnormal processing, unstable mRNA, or non-functional protein.
function of P-bodies
mRNA can be moved to regions of the cytosol called P-bodies (P for Processing)
P-bodies are sites of mRNA degradation or for storing translationally repressed mRNA
Reversibility of P-bodies
mRNA can also be moved back to the cytoplasm to reactivate translation (there is not a membrane around these regions)
function of stress granules
Under conditions of stress, cells accumulate mRNA in another membrane-less compartment called stress granules
In stress granules, translation initiation is blocked until the stressful conditions are removed
Reversibility of p-bodies
mRNAs await return to translation
Enzymes most likely to be present in P-bodies
Deadenylases : remove poly(A) tails
Decapping enzymes : remove 5′ caps from mRNAs
5′→3′ exonuclease: degrades uncapped mRNA
RNA-binding proteins involved in translational repression and decay
RNA interference protein
Enzymes present in stress granules
Translation repressors
Translation initiation factors: involved in stalled initiation
RNA-binding proteins
LncRNA role in regulation
long non-coding RNA molecules that fold into complex structures
Can bind complementary sequences and recruit proteins to act on those genes (usually repressive)
does LncRNA act on dna or rna
Can act on RNA or DNA
Regulation of mRNA by RNA interference
RNA interference (RNAi) involves a short single -stranded RNA binding to a complementary RNA sequence and directing protein machinery to that site
Targeting an mRNA can result in inhibition of translation or mRNA degradation
Binding to an mRNA in the process of being transcribed can result in formation of heterochromatin to block further transcription
3 classes of small noncoding RNA for RNAi
small interfering RNA (siRNA), piwi - interacting RNA (piRNA), and microRNA (miRNA)
Processing of miRNA
At least half of human mRNA are regulated by miRNA!
miRNA precursors are synthesized by RNA pol II
The double-stranded structure of the RNA is cropped and exported to the cytosol where it is cleaved again by the Dicer enzyme
The double-stranded RNA is loaded into the RISC complex where 1 of the strands is degraded, leaving the final single-stranded miRNA
Regulating mRNA with miRNA
• The Argonaute protein is part of the RISC complex
• The miRNA is the guide for directing the RISC complex to the target mRNA
• miRNA positions the target mRNA in the Argonaute active site for cleavage
• After degrading a target mRNA, the RISC/miRNA complex is released to act on another mRNA
when can siRNA and miRNA be used by reserachers
siRNA and miRNA can also be used by researchers to ”knockdown” expression of a target mRNA of interest
Design RNA with complementarity to mRNA of interest → reduce expression.
which types of regulation by non-coding RNA involved in X inactivation
Mammalian X-Inactivation in Females Is Triggered by a lncRNA called Xist
What types of regulatory sequences or proteins might help separate transcriptionally active DNA from heterochromatin that surrounds it?
barriers and insulators
CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
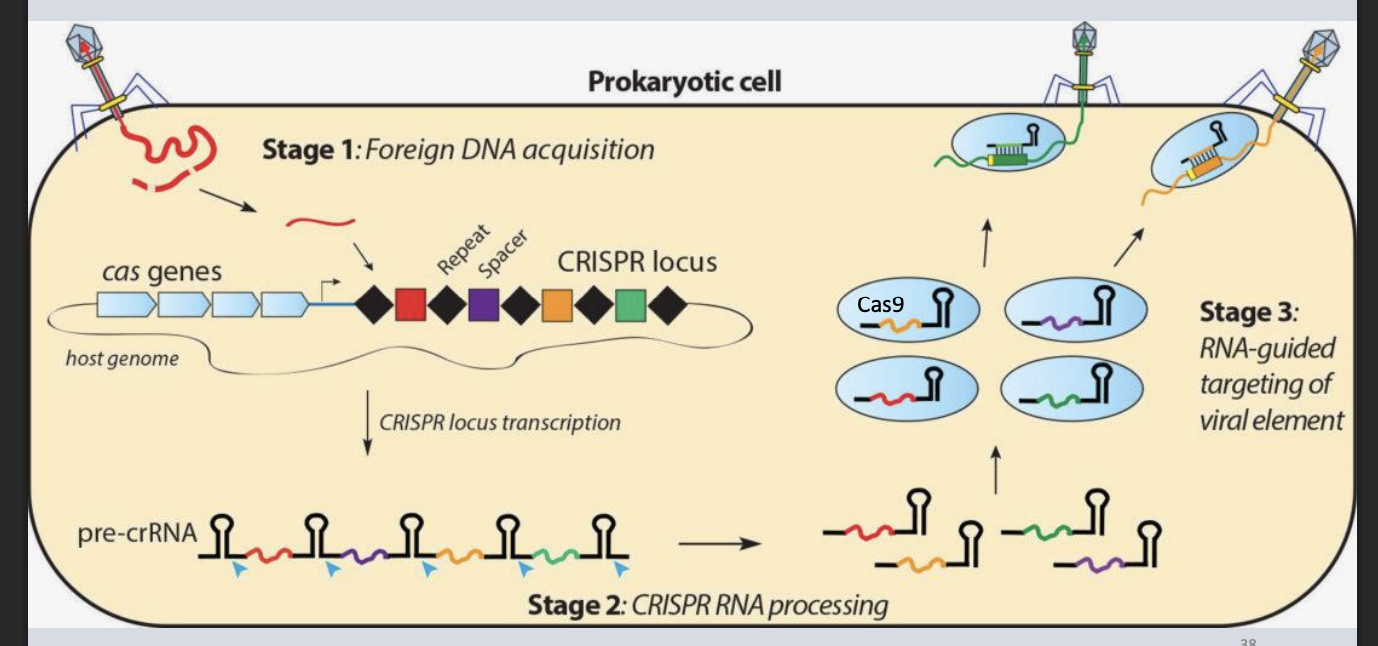
Prokaryotes’s small non-coding RNA defense system
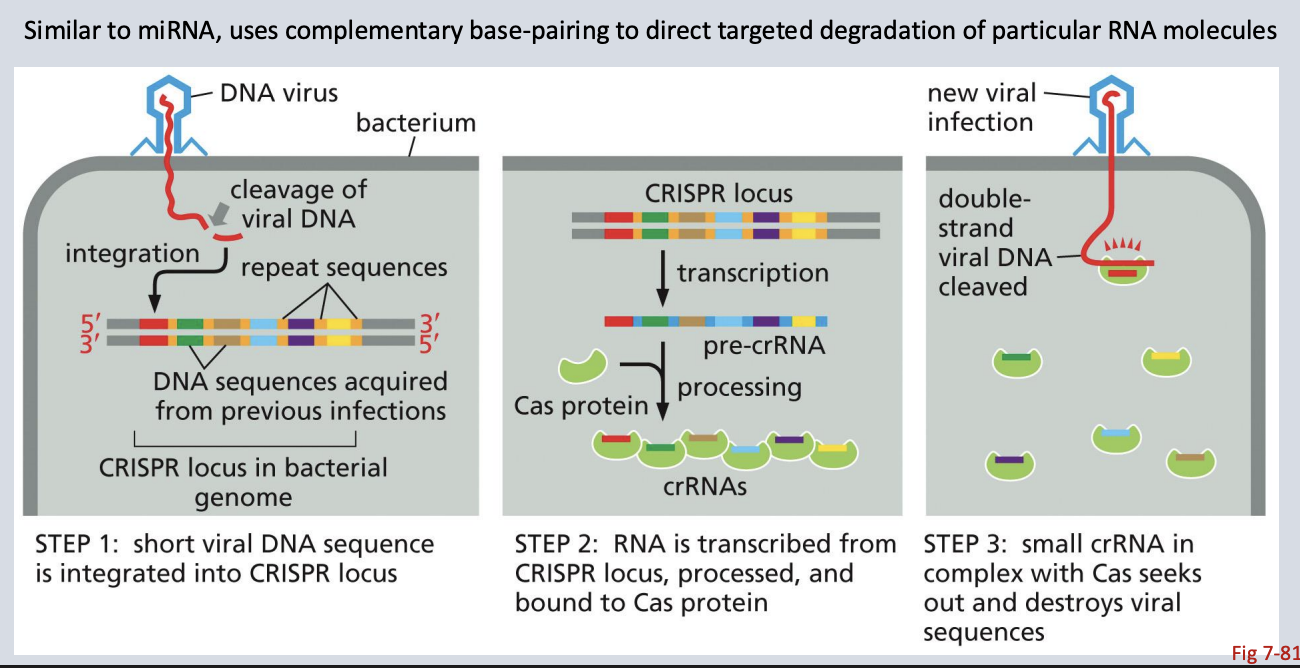
crRNA
Prokaryotic, part of CRISPR-Cas; targets viral RNA/DNA
Describe how crRNA helps protect prokaryotes from viruses
Part of CRISPR system: crRNA guides Cas enzymes to viral DNA.
crRNA binds complementary sequences in invaders and recruits Cas9 to cut them.
Genome editing
can use a gene-specific guide RNA sequence to direct the Cas9 nuclease to a specific site
differentiate how/why homology-directed repair vs NHEJ would be used
Repair of the resulting double-strand break often results in small insertions or deletions (NHEJ)
However, if a plasmid template with homology to the target region is provided to direct repair, then specific changes can be created (Homologous recombination) - ideal for gene therapy
which hr or nhej be preferable for gene therapy approaches in human patients.
For human therapy: HDR is preferable—precise gene editing to fix mutations.
Why might a researcher want to use CRISPR/Cas9 to completely delete a gene from the genome instead of using RNAi to decrease expression?
some small genes have important functions, so sometimes the gene might need to be completely removed
Why might a researcher want to use CRISPR/Cas9 to edit a gene of interest with homology-directed repair?
to make gentic changes to cure diseases make cell and edit them for a good functional cause
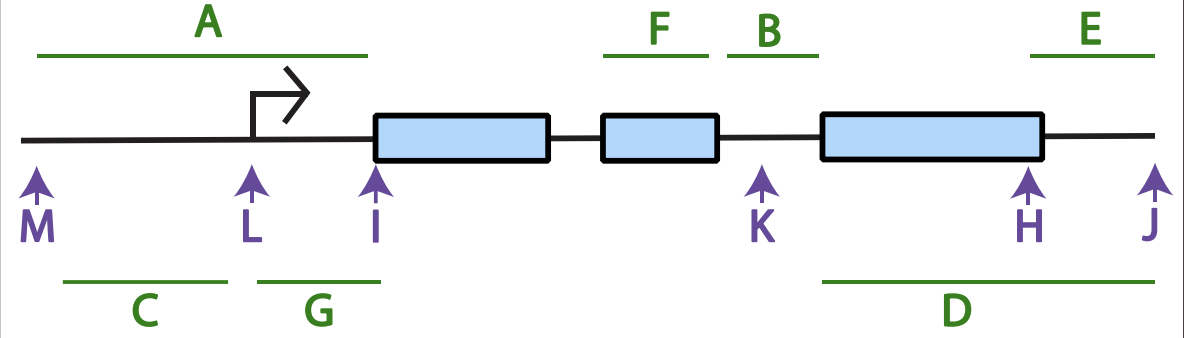
what is the black arroe
are typically used to label where an RNA transcript will begin
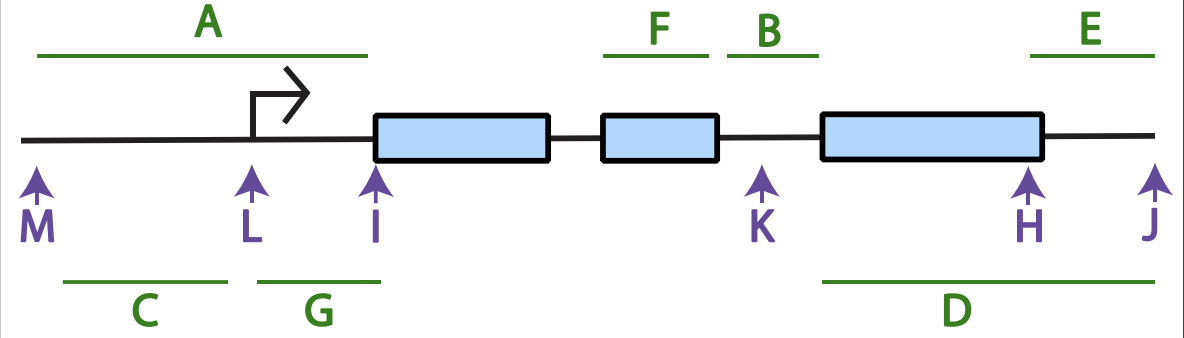
(green letters) Intron:___, promoter:___, 3’ UTR:___, 5’ UTR:___,
Intron: f,
promoter: a,
3’ UTR: e,
5’ UTR: g,
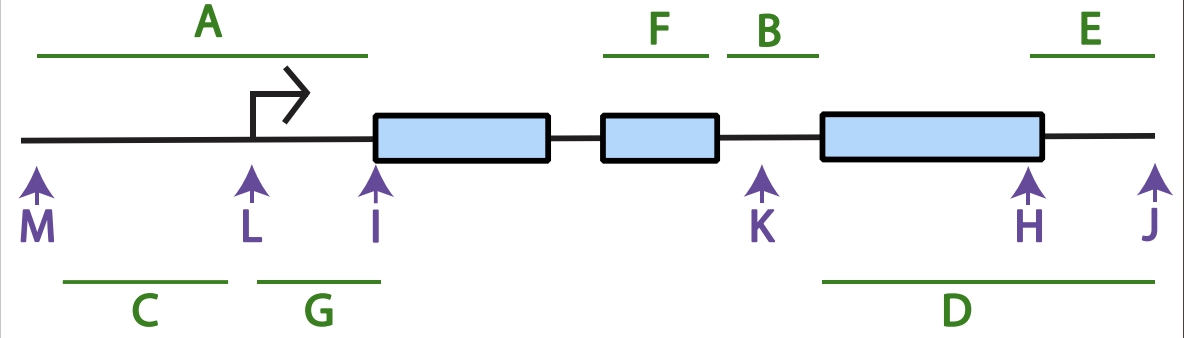
(purple letters) transcription initiation:___, transcription termination:___, start codon:___, stop codon:___, Kozak sequence:___
transcription initiation: L
transcription termination: J
start codon: I
stop codon: H
Kozak sequence:M
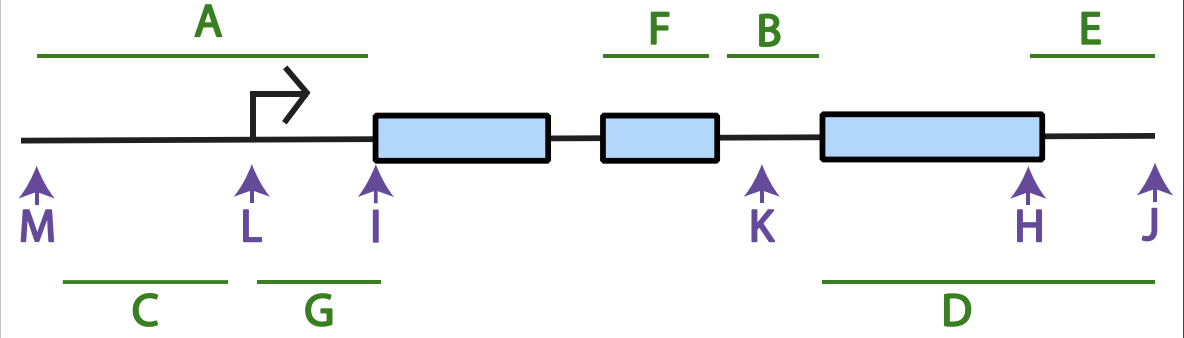
Which letter best corresponds to where polyadenylation will occur on the resulting mRNA?
J
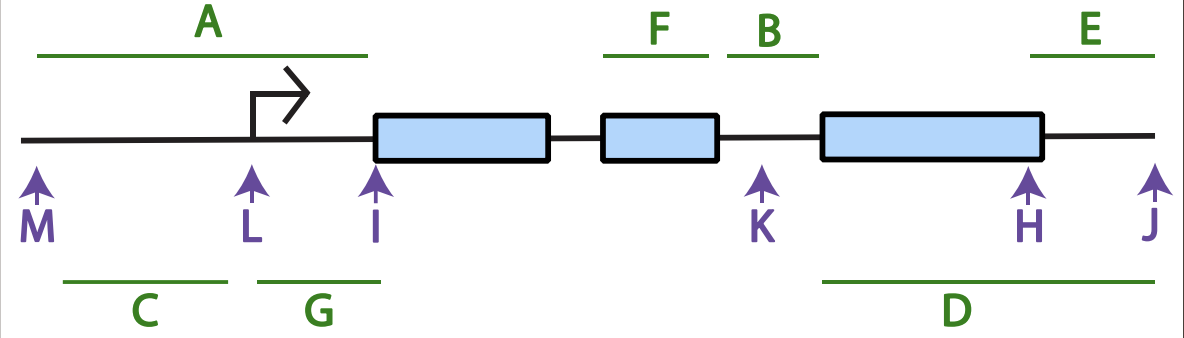
Which letter best corresponds to sequence encoding the N-terminus of protein?
F
Where could an enhancer or silencer sequence be found?
C or A