lec 4 (mcbride) - protein turnover and AA catabolism
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
50 Terms
maintaining homeostasis…
requires metabolic regulation that coordinates the use of nutrient pools
protein digestion and turnover
AA are obtained from diet when proteins are digested
cellular proteins are degraded to AA b/c of damage, misfolding, or changing metabolic demands
excess AA CANNOT be stored or excreted → must be used as metabolic fuel
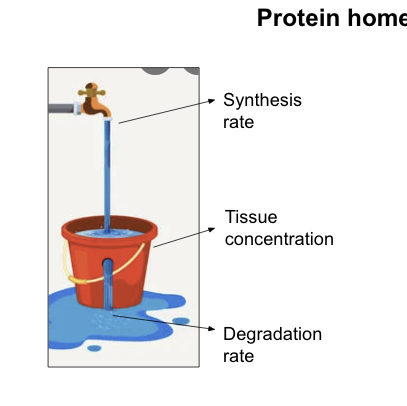
protein production and consumption rates…
are controlled to maintain physiological levels and function
homeostatic mechanisms adjust these rates to achieve production = consumption to maintain a physiological concentration required for life
proteins are degraded to…
amino acids
dietary proteins are degraded to AA which are absorbed by the intestine and transported in the blood
essential AA = AA that CANNOT be synthesized and must be acquired in the diet
essential AA
in humans CANNOT be synthesized from other dietary precursors
histidine
isoleucine
leucine
lysine
methionine
phenylalanine
threonine
tyrptophan
valine
digestion of dietary proteins
begins in the stomach and is completed in the intestine
protein digestion begins in the stomach where the acidic environment denatures proteins into random coils
pepsin = the primary proteolytic enyzme of the stomach
max active at pH 2
partly digested proteins move from the stomach → beginning of the small intestine (duodenum) → stimulating secretion of sodium bicarbonate and proteolytic enzymes from the pancreas
aminopeptidases in the plasma membrane of intestinal cells enhance digestion
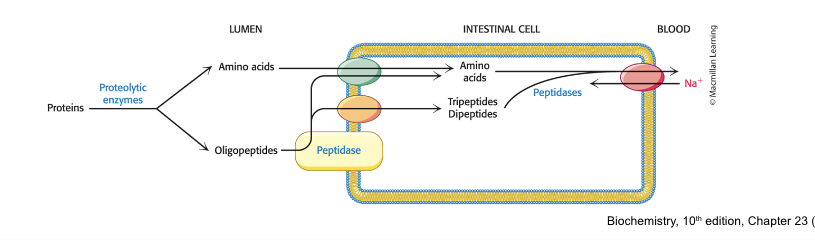
products of protein digestion…
are absorbed by the small intestine
free AA, dipeptides, and tripeptides are transported into the intestinal cells
at least 7 different transporters exist, each specific to a different group of AA
absorbed AA are released into the blood by a number of Na+-AA antiporters
cellular proteins are degraded at…
different rates
protein turnover = the degradation and resynthesis of proteins
takes place constantly in cells
essential for removing short-lived or damaged proteins
the half-lives of proteins range over several orders of magnitude
ornithine decarboxylase, which catalyzes the synthesis of poly amines, is ~11 mins
hemoglobin = life of RBC
lens protein crystallin = life of organism
protein turnover
is tightly regulated
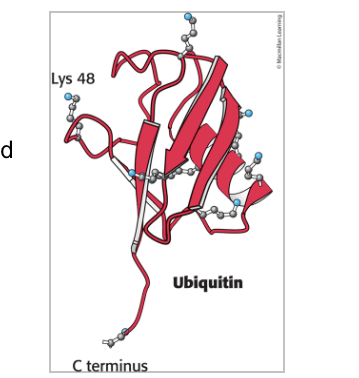
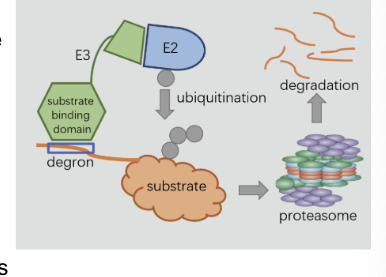
ubiquitin = a small (76 aa) that tags proteins for destruction
present in all eukaryotic cells
highly conserved
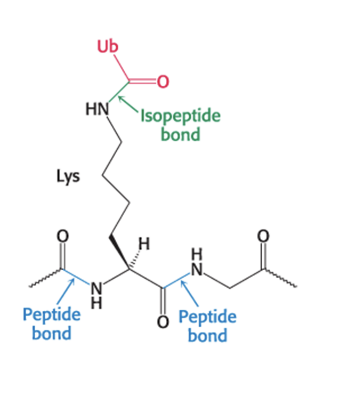
ubiquitin attaches by the carboxyl-terminal Gly residue to the ε-amino groups of +1 lys residues on target protein
requires ATP hydrolysis
forms an isopeptide bond because ε rather than α-amino groups are targeted
ubiquitin
is a small, compact protein with 7 lysine residues
has an extended carboxyl terminus which is activated and linked to proteins targeted for destruction
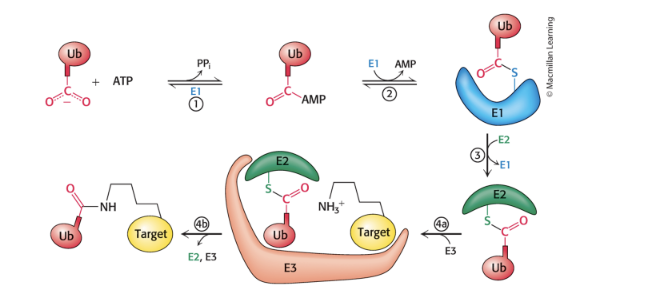
3 enzymes participate in the attachment of ubiquitin to protein
ubiquitin-activating enzyme (E1) = adenylates ubiquitin and transfers it to a sulfhydryl group of a Cys residue of E1
ubiquitin-conjugating enzyme (E2) = transfer ubiquitin to one of its own sulfhydryl groups
ubiquitin-protein ligase (E3) = transfers ubiquitin from E2 to an ε-amino group of target protein
brings E2 and target protein together
ubiquitin be transffered directly or be passed to a cys residue of E3 first
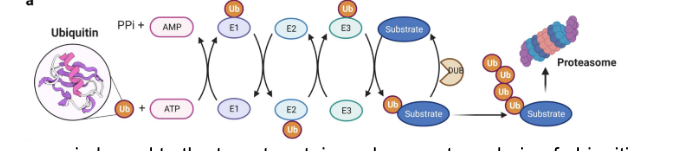
multiple ubiquitin molecules are typically…
added to a single protein substrate
E3 can remain bound to target protein and generate a chain of ubiquitin molecules
E3 can dissociate after the first ubiquitin addition and a chain can be extended by another E2/E3 pair
ubiquitin can be added onto any of the 7 Lys or the N-terminus of the previous ubiquitin
a chain of 4+ ubiquitin molecules linked via Lys 48 = especially effective signal for protein degradation
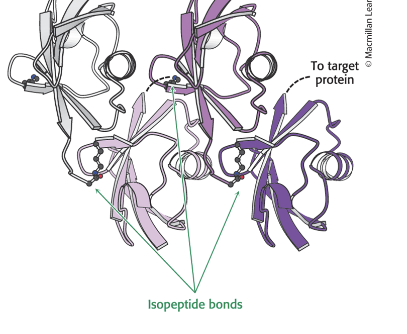
E3 ubiquitin ligases provide…
the protein target specificity
in humans:
2 E1 ligase genes
30-50 E2 ligase genes
over 600 E3 ligase genes
in a tetraubiquitin chain…
4 ubiquitin molecules are linked by isopeptide bonds
the ε-amino group of a Lys residue of 1 ubiquitin is linked to the terminal carboxylate of another
this unit = primary signal for degradation when linked to target protein
degrons
are amino acid sequences that control protein half lifew
degron = specific sequence of AA that indicate a protein should be degraded
for many proteins, the amino-terminal residue AA (N-degron) = important degradation signal for E3 enzyme
may only be exposed after proteolytic cleavage
may be added after protein synthesis
may require other modifications
other degrons include cyclin destruction boxes and PEST sequences
cytoplasmic yeast
dependence of the half-lives of cytoplasmic yeast proteins on the identity of their amino-terminal residues
highly stabilizing residues (half life > 20 hours)
ala
cys
gly
met
pro
ser
thr
val
intrinsically destabilizing residues (half life = 2-30 mins)
arg
his
lie
leu
lys
phe
trp
tyr
destabilizing resides after chemical modification (half life = 3-30 mins)
asn
asp
gln
glu
importance of E3 proteins to normal cell function
proteins that are NOT degraded b/c of a defective E3 may accumulate → causing a disease of protein aggregation
angelman syndrome = severe neurological disorder characterized by an unusually happy disposition, cognitive disability, absence of speech, uncoordinated movement, and hyperactivity
caused a defect in a member of E3 family
overexpression of E3 ligase causes autism
inappropriate protein turnover → cancer
additional roles of ubiquitination
ubiquitination also regulates proteins involved in:
DNA repair
chromatin remodeling
innate immunity
membrane trafficking
autophagy
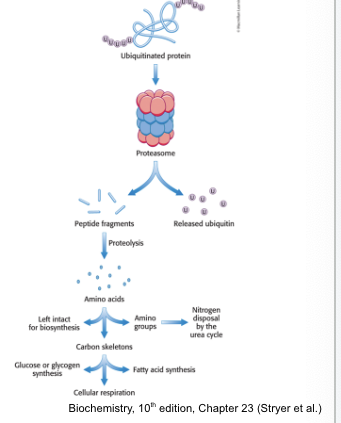
the proteasome
digests the ubiquitin-tagged proteins
proteasome (26S proteasome) = a large, ATP-driven protease complex that digests ubiquitinated proteins
the 26S proteasome = complex of 2 components
one 20S catalytic unit arranged as barrel
two 19S regulatory units that control access to the interior of the 20S catalytic subunit
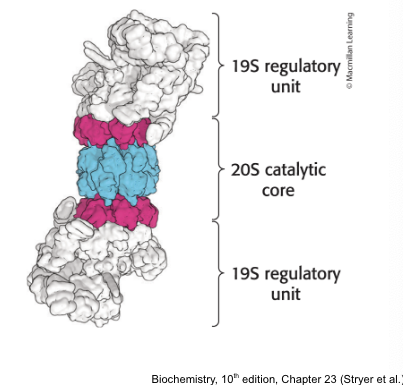
functions of the 19S regulatory unit
the 19S regulatory units:
contain ubiquitin receptors that bind specifically to polyubiquitin chains
uses ATP to unfold polyubiquitinated chains and direct them into catalytic core
contains an isopeptidase that cleaves off intact ubiquitin molecules so they can be reused
key components of the 19S complex = 7 ATPases of the AAA+ class
a class of chaperone-like ATPases associated with:
assembly
operation
and disassembly of protein complexes
20S proteasome
is barrel-shaped and made up of 28 homologous subunits
the subunits (α-type, red; β-type, blue) are arranged in 4 rings of 7 subunits each
some of the β-type subunits (right) include protease active sites at their amino termini
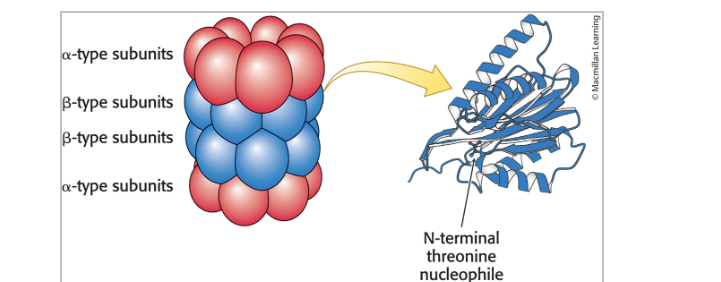
proteolytic active sites of the 20S barrel
there are 3 types of active sites in the β subunit, each with different specificity
chymotrypsin-like: cleaves after large hydrophobic amino acids
trypsin-like: cleaves after basic amino acids
caspase-like: cleaves after acidic amino acids
all active sites employ an N-terminal Thr residue
the OH group of the Thr residue attacks the carbonyl groups of peptide bonds → forms acyl-enzyme intermediates
substrates are degraded in a processive manner WITHOUT intermediate release
substrates are reduced to peptides ranging from 7-9 residues before release
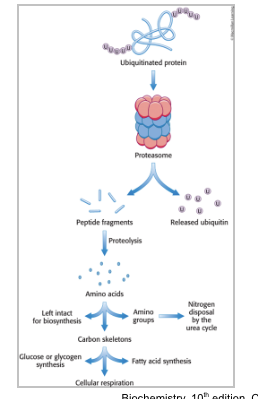
the proteasome and other proteases generate
free amino acids
ubiquitinated proteins are processed to peptide fragments
ubiquitin is removed and recycled prior to protein degradation
peptide fragments are further digested to yield free AA which can be used for biosynthetic reactions; most notably protein synthesis
alternatively, the amino group can be removed and processes to urea and the carbon skeleton can be used to synthesize carbohydrate or fats or used directly as fuel for cellular respiration
processes regulated by protein degradation
many biological processes are controlled, at least in part, by protein degradation via the ubiquitin-proteasome pathway
processes regulated by protein degradation
gene transcription
cell-cycle progression
organ formation
inflammatory response
tumor suppression
cholesterol metabolism
antigen processing
protein degradation can be used to regulate…
biological function
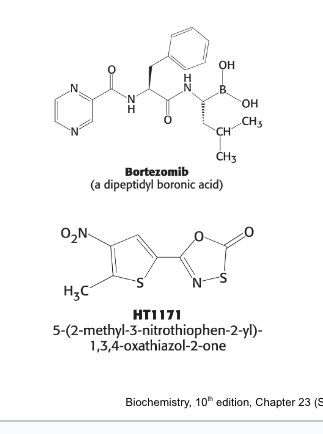
bortezomib (velcade) = dipeptidyl boronic acid inhibitor of the proteasome
used as therapy for multiple myeloma
degrons = used as regulatory mechanisms for protein expression
HT1171 = suicide inhibitor of the proteasome of M. tuberculosis
has NO effect on human proteasomes
first step in amino acid degradation
is the removal of nitrogen
AA NOT needed as building blocks are degraded to compounds able to enter the metabolic mainstream
amino group is removed and remaining carbon skeleton is metabolized to glycolytic intermediate or to acetyl CoA
major site of AA degradation in mammals = liver
muscles also readily degrade the branched-chain AAs (leu, Ile and Val)
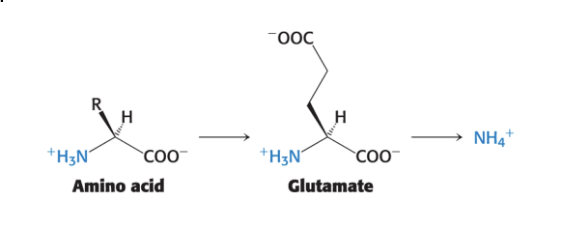
alpha-amino groups
are converted into ammonium ions by oxidative deamination of glutamate in liver
α-amino groups → α-ketoglutarate → yielding glutamate
glutamate is oxidatively deaminated in the liver to yield ammonium ion (NH4+)
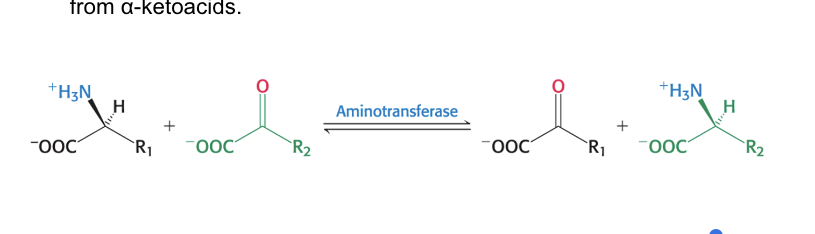
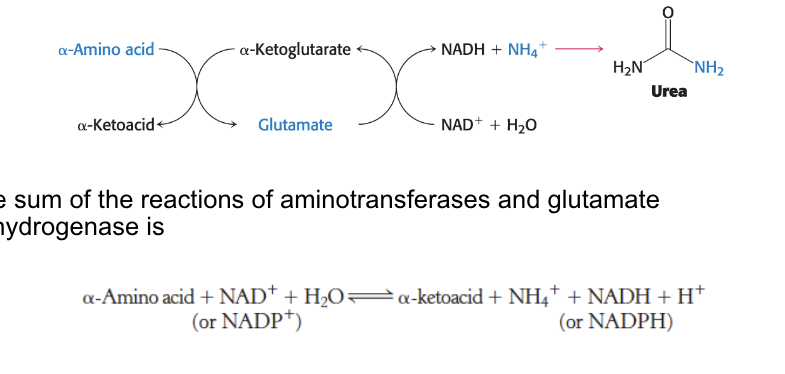
role of aminotransferases
aminotransferases (transminases) = catalyze the transfer of α-amino group from an α-amino acid → α-ketoacid
reaction are reversible and can be used to synthesize amino acids from α-ketoacids
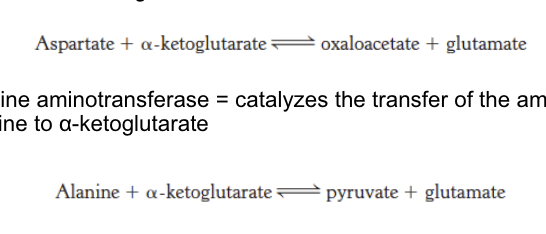
aspartate aminotransferase and alanine aminotransferase
aspartate aminotransferase = catalyze the transfer of amino group of aspartate → α-ketoglutarate; end result = oxaloacetate + glutamate
alanine aminotransferase = catalyze the transfer of the amino group of alanine → α-ketoglutarate; end result = pyruvate + glutamate
blood levels of aminotransferase serve as…
diagnostic function for liver damage
the presence of alanine and aspartate aminotransferase in the blood = indication of liver damage
liver damage can occur due to:
viral hepatitis
long-term excessive alcohol consumption
reaction to drugs
in cases of liver damage, liver cell membranes are damaged and aminotransferases leak into blood
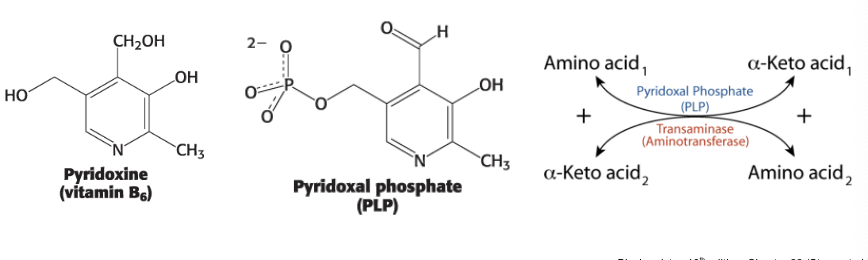
aminotransferases require…
pyridoxal phosphate (PLP) from vitamin B6
aminotransferases require the coenzyme pyridoxal phosphate (PLP) - a derivative of pyridoxine (vitamin B6)
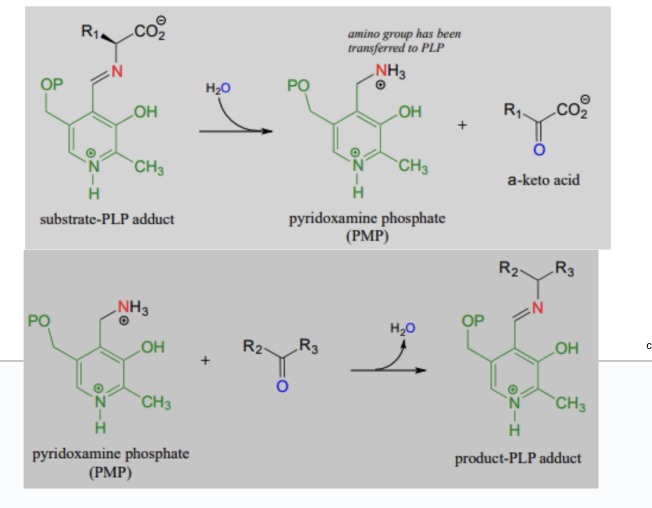
transamination
step 1
transfer of amino acid group from amino acid substrate → PLP and release of keto acid
example: for alanine aminotransferase, AA = alanine and released ketoacid is pyruvate
step 2
transfer of amino acid from coenzyme → keto acid → new amino acid
example: for alanine aminotransferase, the ketoacid is α-ketoglutarate and the released AA is glutamate
pyridoxal phosphate enzymes
catalyze a wide array of reactions
at the α-carbon of amino acids, PLP-dependent enzymes catalyze:
decarboxylations
deaminations
racemizations
aldol clevages
at the β-carbon and γ-carbon of AA, PLP-dependent enzymes catalyze elimination and replacement rxn
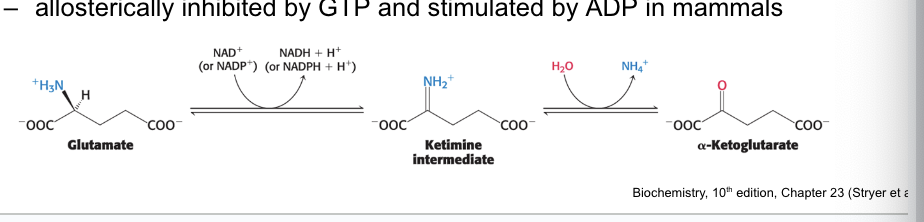
role of glutamate dehydrogenase
glutamate dehydrogenase = a mitochondrial enzyme that converts the nitrogen atom in glutamate → free ammonia ion by oxidative deamination
is essentially a liver-specific enzyme
can use either NAD+ or NADP+
proceeds by dehydrogenation of the C-N bond, followed by hydrolysis of the ketimine
allosterically inhibited by GTP and stimulated by ADP in mammals
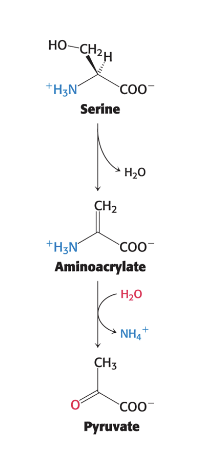
serine and threonine can be…
directly deaminated
serine dehydratase and threonine dehydratase directly deaminate their respective AA
PLP = prosthetic group (non-protein component that is tightly bound to protein and essential for biological function)
NO transfer of the α-amino group to α-ketoglutarate is required
dehydration precedes deamination
serine → pyruvate + NH4+
threonine → α-ketobutyrate + NH4+
fate of the ammonia ion
in most terrestrial vertebrates, NH4+ converted into urea → excreted
sum of the reactions of aminotransferases and glutamate dehydrogenase is → second equation in image
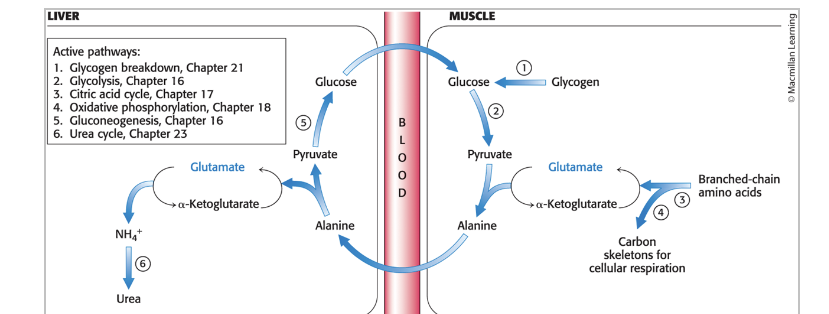
peripheral tissues transport…
nitrogen to the liver
muscles use branched-chain AA as fuel during prolonged exercise and fasting
muscles lack enzymes of the urea cycle
nitrogen is transported from muscle → liver as alanine (through glutamate) in the glucose-alanine cycle
glutamine synthetase = catalyzes the synthesis of glutamine from glutamate and NH4+
nitrogens of glutamine can be eliminated by incorporation into urea in the liver

pathway integration: the glucose-alanine cycle
allows muscle cells to use AA as fuel
during prolonger exercise and fasting, muscles use branched-chain AA as fuel
nitrogen removed is transferred (through glutamate) to alanine which is released into the blood stream
in the liver, alanine is taken up and converted into pyruvate for the subsequent synthesis of glucose
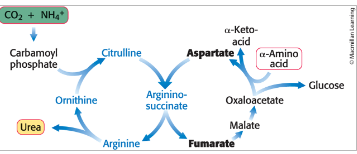
urea cycle
eliminates both nitrogen and carbon waste products
2 nitrogen atoms enter the cycle and leave as urea
carbon dioxide is simultaneously eliminated as it is hydrated to bicarbonate which then enters the cycle
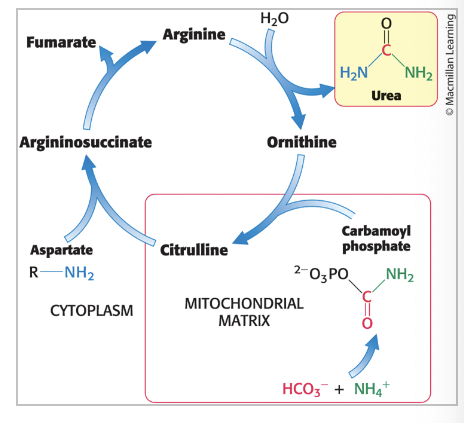
urea cycle begins with…
formation of carbamoyl phosphate
carbamoyl phosphate synthetase I = catalyzes the coupling of ammonia (NH3) with bicarbonate (HCO3-) → form carbamoyl phosphate
occurs in mitochondria
mammals have 2 isozymes
requires 2 molecules of ATP, making reaction essentially irreversible
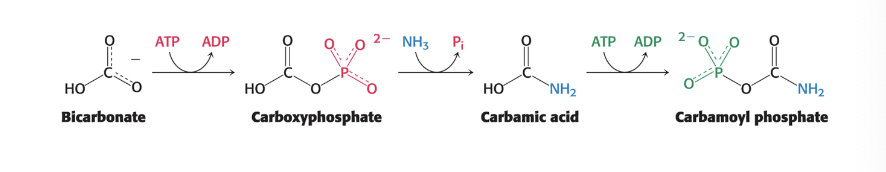
carbamoyl phosphate synthetase I is…
the key regulatory enzyme for urea synthesis
carbamoyl phosphate synthetase I:
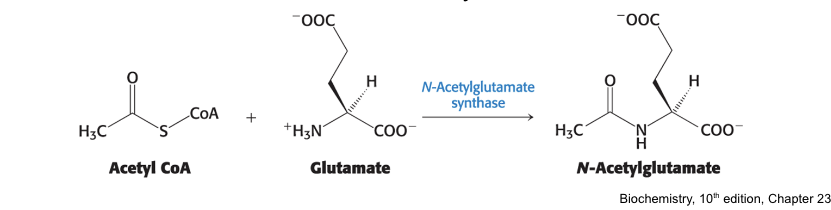
requires the allosteric regulator N-acetylglutamate for activity
is inhibited by acetylation and stimulated by deactylation
N-acetylglutamate synthase = catalyzes the synthesis of N-acectylglutamate
activated when AA are readily available
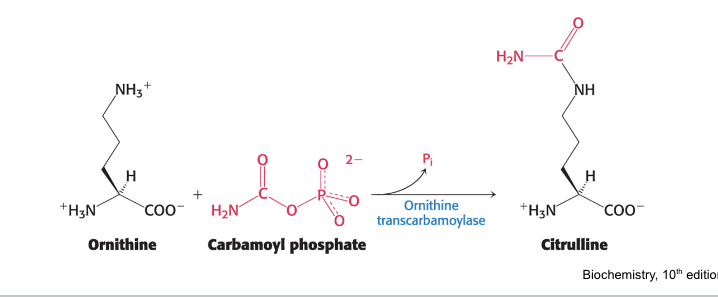
carbamoyl phosphate reacts with…
ornithine to begin urea cycle
ornithine transcarbamoylase = catalyzes the transfer of the carbamoyl group of carbamoyl phosphate to ornithine forming citrulline
occurs in the mitochondria
citrulline is transported into the cytoplasm
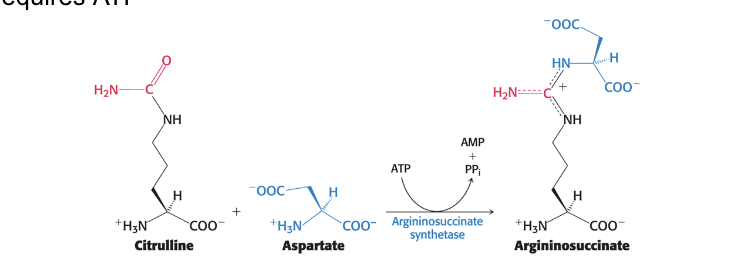
citrulline
condenses with aspartate
aspartate is the donor of the second nitrogen of urea
argininosuccinate synthetase = catalyzes the condensation of citrulline and aspartate → argininosuccinate
occurs in cytoplasm
requires ATP
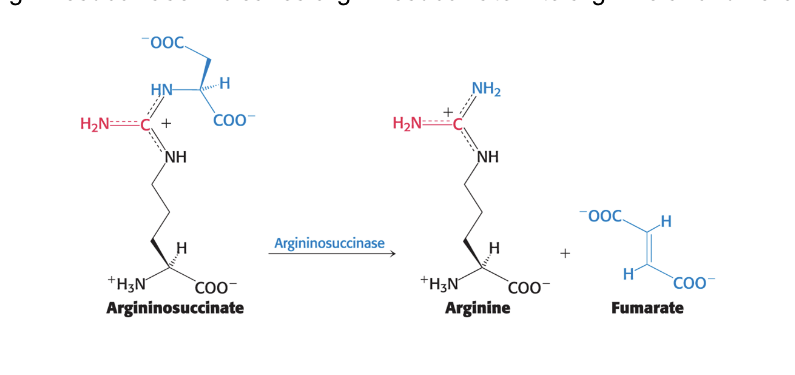
cleavage of argininosuccinate
argininosuccinase = cleaves argininosuccinate → arginine and fumarate
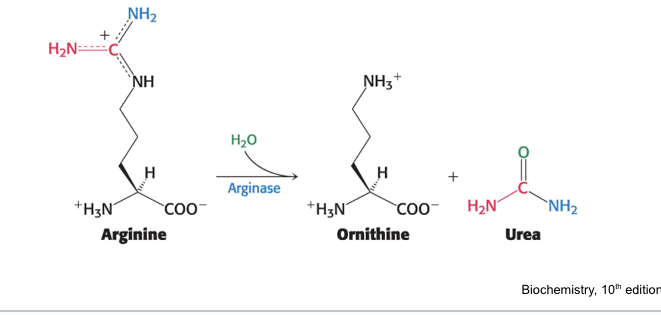
hydrolysis of arginine
arginase = hydrolyzes arginine to generate urea and ornithine
ornithine is transported back into mitochondria
urea is excreted
urea cycle is linked to gluconeogenesis
the stoichiometry of the urea cycle = photo
fumarate is hydrated to malate which is in turn oxidized oxaloacetate
oxaloacetate can be:
converted into glucose by gluconeogenesis
transaminated to aspartate for another round of urea synthesis
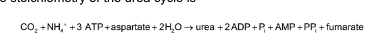
nitrogen metabolism is…
integrated with other metabolic pathways
urea cycle, gluconeogenesis, and the transamination of oxaloacetate are linked by fumarate and aspartate
inherited defects of the urea cycle…
cause hyperammonemia and can lead to brain damage
any defect in the urea cycle leads to an elevated level of NH4+ in the blood (hyperammonemia)
high levels of NH4+ may:
inappropriately activate an Na+-K+-Cl- cotransporter, disrupting the osmotic balance of the nerve cell and causing cellular swelling
disrupt neurotransmitter systems
impact energy metabolism, levels of oxidative stress, nitric oxide synthesis and signal transduction pathways
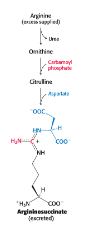
argininosuccinase deficiency
can be managed by supplementing the diet with arginine
argininosuccinate deficiency is treated by:
restricting total protein intake
supplementing the diet with arginine
excess nitrogen is excreted in the form of argininosuccinate
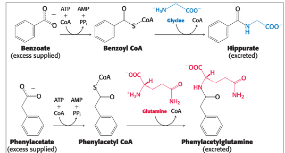
both carbamoyl phosphate synthetase and ornithine transcarbamylase deficiencies…
can be treated with supplementation
by supplementing the diet with benzoate and phenylacetate, excess nitrogen can be excreted in the form of hippurate and phenylacetylglutamine