Transcriptional Activation, Chromatin Modifications, and Gene Regulation in Eukaryotes
1/99
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
100 Terms
the job of a transcriptional activator is to...
get all the machinery to DNA (mediator, polymerase, and all of the Ts) to the right place to transcribe the gene
sequence specific DNA binding...
helps to bring all the machinery to transcribe the gene
does phosphorylation increase or decrease interaction between proteins?
we only know it changes but we don't know in which direction
Because CREb is always bound to DNA which we know from CHIP but is activated when phosphorylated...
there is something that binds to phosphorylated CREB or is free from phosphorylated CREB that is triggering teh activation of genes
How does phosphorylated CREB activate trasncription?
1. took recombinant CREB and phosphorylated it with PKA.
2. Then they used that as a probe to screen an "expression" library
3. that led to CBP (CREB-Binding Protein) to be identified
4. this led to questions:
the CBP binding to phosphorylated CREB led to questions:
a) is CBP binding to CREB-PKA dependent? (this is important if this is the way that CREB activates genes)
b) is CBP binding to CREB-target genes PKA-dependent?
c) what does CBP do?
how do you test: Is CBP binding to CREB-PKA dependent? (CBP-CREB direct interaction)
GST
1. we're going to take some GST-CREB AD fusion protein and add it to two tubes
2. We're going to add PKA to one tube the other we'll be left unphosphorylated (we would do this by incubating GST-CREB with PKA with magnesium and ATP)
3. Wash PKA after it has been phosphorylated. Now we have purified GST-CREB
4. To the third tube and fourth tube we add GST -CREB AD with mutated ser- ala (point mutation)
5. incubate one with PKA to attempt to phosphorylate it.
6. do not add PKA to the other
7. We should not see an interaction between the CREB and CBP because CREB cannot be phosphorylated . Therefore, we are able to say that interaction is solely between CBP and CREB
8. Wash away PKA
9. Use radio labeled CBP whcih will bind only the GST_CREB AD (normal) that was incubated with PKA
10. add glut beads to pull GST down
11. wash away supernatant
12. Add SDS and run denatured proteins on a gel
how do you test: is CBP binding to CREB-target genes PKA-dependent? (CBP recruitment to CREB-regulated DNA)
chIP:
1. Cross links will be formed between CREB and DNA and CBP and CREB
2. Use and antibody to CBP
3. IMMunoprecipitate DNA that is bound to CREB
4. Identify direct and indirect interactions
5. IT was found that there was very little CBP binding to CREB in low PKA whereas there were a lot of interaction is high PKA
how do you test: what does CBP do?
1. Sequence the gene and look for homology to other proteins
CBP is also called a...
coactivator because it is being recruited to DNA by many different trasncriptional activator domains
CBP binds to...
CREB and c-fos
is bringing CBP to a gene sufficient to activate trasncription?
experiment:
1. Reporter gene with: 5 UAS---USF bs---- TATA---Initiator---Reporter gene
2. UAS sites are for the GAL 4 DBDn to bind
3. First tested GAL-4 on its own to see if that would lead to trasncription of reporter gene
4. Then they did a Gal 4 DBD - CBP HAT fusion protein to see if it activates trasncription
5. It was found that there was 10x more reporter gene activity with teh fusion protein rather than just gal 4 alone
5. the HAT domain of CBP is sufficient to activate trasncription
6. Control: Gal 4 DBD- CBP HAT mutated which inactivates the HAT domain which led to even less trasncription of the reporter gene
what is HAT...
Histone acetyl transferase- enzyme that will transfer an acetal group to a histone
- it is a version of a "writer" because it adds an acetal group to a histone tail
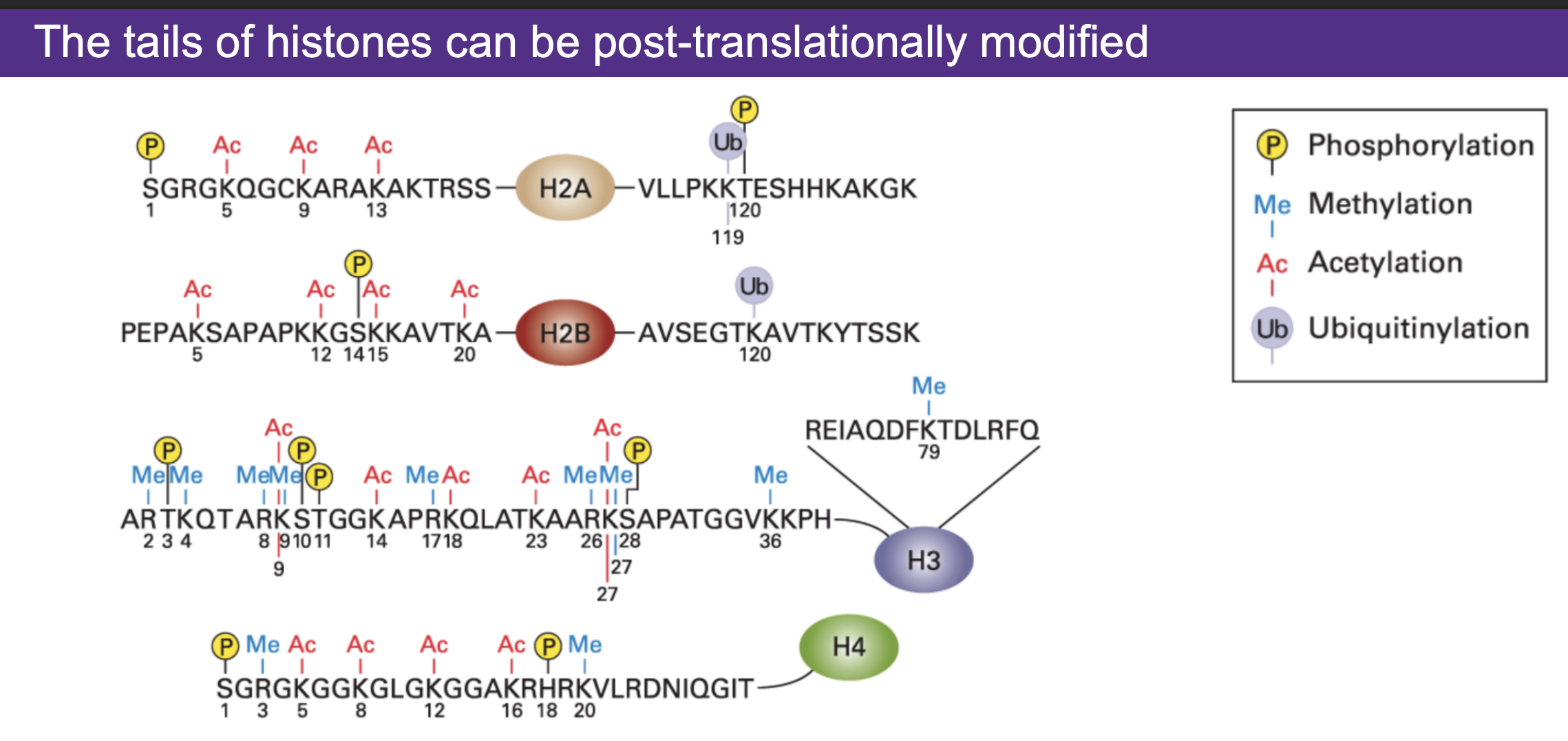
The tails of histones can be post-translationally modified...
1. ADD; writer
2. DELETE: eraser
Nuclosomes and histones
1. DNA is wrapped around nucleosomes in the form of an octamer with 2 H3, 2 H2A, 2 H2B, 2 H4 histones
2. Histones have tails that can get modified
3. Some enzymes will add a modification: "writers"
4. Some enzymes remove modification: "eraser"
5. readers: read the state of the histone tail
modifications of histone tails are:
1. phosphorylation
2. methylation
3. acetylation
4. ubiquitinylation
modifications of histones can be at...
the N terminus and C terminus of histones
Which amino acid is most commonly modified in histones?
K which stands for Lysine
What is special about Lysine in the context of DNA?
1. Lysine is basic and positively charged whereas DNA is negatively which means that Lysine can bind DNA.
2. histones are rich in lysine so they bind DNA tightly
Histones have what type of post-translational modification
bi-directional
Bi-directional post-translational modification
1. transcriptional activators recruit HAT: leads to acetylated histone K9 or K27
2. trasncriptional repressors recruit HDAC (histone deacetylase): removes acetal group from histone
Modifications of histones are examples of...
epigenetics
if chromatin is totally open...
it is easier for trasncription factors to bind there and help recruit polymerase
if chromatin is condensed in heterochromatin form...
it's going to be hard for a transcription factor to bind DNA and even harder for RNA polymerase to get in there and transcribe a gene
when you add a HAT, it makes chromatin...
more open for easier transcription
when you add HDAC, it makes chromatin...
more condensed and harder to be transcribed
acetylation is associated with...
active trasncription
methylation is associated with...
repressors which leads to a repressed state of chromatin
HDAC
histone deacetylase
- removes acetal group from histone
HDMT
histone demethylase
- moves histone towards the demethylated state by removing methyls from K9 and K27
HMT
histone methylase
- adds methyls to K9 and K27
when you add a HDMT, it makes chromatin...
it demethylates chromatin it makes chromatin more open
when you add a HMT, it makes chromatin...
makes histones methylated and more condensed
Methylation compacts gene...
- enzyme methyl trasnferase methylates Histone H3 K9 and/or K27
- that methylated histone H3 is a signal for one of the readers to come and bind it
- this reader is known as HP1
- HP1 binds to methylated Histone H3
- Then you get an oligomerization of HP1 reader proteins with the chromatin
- this draws all the DNA together and you get very condensed DNA
HP1
Heterochromatin Protein 1
Once the HP1 binds the methylated K9 and or K27 as a case of the repressor methyltransferase...
the methyltrasnferase repressor will degrade over time but the chromatin will remain in condensed form for way longer... you can fairly permanent inactivation of a part of a genome
Using chip-seq to analyze distribution of a gene we would see CBP?
peaks where CRE is (where CREB binds) only
Using chip-seq to analyze distribution of a gene we would see H3 K27 ac?
across the promoter region and across the trasncribed region (all of it) this indicates that there is a lot of acetylation
Using chip-seq to analyze distribution of a gene we would see H3 K9 Me?
across the whole DNA (and everything else is low)
What would be somethimg that you could chip teh region of the genome to show that it is in heterochromatin form?
antibody against HP1
You can use chip-seq with acetylaytes or methylated histones..
to get a sense of what genes are being trasncribed then do RNA sequencing of those cells
c-fos is a...
trasncription factor
in a fibroblast the Creb, c-fos, fos pathway...
leads to cell cycle entry
in neurons, neural firing has teh response of...
CREB getting phosphorylated in response to neurons firing action potentials but ultimately leads to neuronal plasticity from FOS
why is it important to not induce cell cylce entry in neurons?
because there are no new neurons so it is imprtnat that they do not enter a cell cycle
How can the same TF (c-fos) activate different genes in different cells?
Chromatin
- some genes will lead to cell cycle entry
- some genes will lead to neuronal plasticity
A neuron: will put all cell cycle entry genes into a heterochromatin state to not allow FOS to bind
A fibroblast: will condense all genes involved in neuronal plasticity to not allow FOS to bind to those and only to cell cylce entry
How are eukaryotic trasncription factors regulated?
1. Regulating nuclear localization
2. Phosphorylation to activate TF
3. Increase gene expression
4. Repressors
5. Chromatin
Pioneer TFS hold genes...
open for transcription like GATA 3 which holds DNA in an open conformation
Alternative splicing real world diagram
arrow shows to direction of trasncription
Alternative splicing real word diagram how to determine transcription start site?
Check to see if non coding boxes in the 5'UTR region line up
Alternative splicing real word diagram how to determine RNA isoforms?
check if boxes are all the same
Alternative splicing real word diagram how to determine protein isoforms?
look at coding boxes
what is the different between these isoforms? what is the orgnaism going to gain from having so many RNA isoforms?
The difference is in the UTR meaning that there must be information in the 5 and 3 UTR
If an RNA isoforms only have one transcription start site (no difference across the isoforms) only, there is only one protein produced (because all teh coding regions are the same, and all the 3' UTRs are the same...
the 5' UTR is what is different and some regulation in the 5'UTR
when an gene has many different isoforms..
it doesn't mean that every cell in teh body is going to make all the different forms of RNA and proteins, some isoforms will be made in one art of the body and others in another part. Each of all the isoforms have different forms of regulation
So you can take one gene and by expressing it a different levels and with slightly diffenret versiond of the proteins with slightly differnet 5'UTR and 3'UTR...
you can actually diversify what you've got
what is lncRNA?
long non coding RNA and it is included in some of the 3'UTR of some isoforms but we don't understand what it does. we know it doenst code for a protein but it will be trasnribed in specific cells
Alternative splicing in human genes
95% of genes with exons undergo alternative splicing
Diseases and cancer are associated with alternative splicing...
15% of human hereditary diseases
Because of alternative splicing, we can go from 20,000 human genes
to around 130,000 different proteins
the gene in Drosophila called Dscam encodes...
cell adhesion molecule and helps neurons stick to each other
how many possible Dscam isoforms
12x48x33x2= 38016
Alternative splicing: drosophila sex determination, in females... (don't need to remember this perfectly)
1. Sxl is on the X chromosome and because female droshopila flies have one more chromosome than males, they have increased levels of Sxl
2. Sxl protein binds to sxl RNA and leads to splicing out exon 3 and a 2-4 mRNA which codes for more Sxl protein
3. Sxl protein binds to tra RNA and leads to splicing out of exo 2 (which has a stop codon) and a 1-3 mRNA which codes for Tra protein
4. Tra protein binds to dsx RNA and leads to splicing 3-4 mRNA (4 has polyadenylation)
5. the 3-4 mRNA codes for DsxF protein which leads to female gene expression
Alternative splicing: drosophila sex determination, in males... (don't need to remember this perfectly)
1. With lower levels of Sxl because they only have on X chromosome, the Sxl is not alternatively spliced and the mRNA ends up being 2-3-4. Exon 3 has a stop codon and the mRNA does not code for Sxl protein
2. Without the Sxl protein, teh tra RNA is not alternatively spliced and the mRNA ends up being 1-2-3. Exon 2 has a stop codon and the mRNA does not code for Tra protein
3. Without Tra protein, dsx RNA is alternatively spliced and the mRNA produced is 3-5. The mRNA codes for Dsx M protein which leads to male expression
Sxl and Tra protein
are proteins that regulate splicing
What fluorescent protein can be used for an imaging experiment where you are looking at DNA?
a fusion protein with histone H2B and BFP (blue fluorescent protein) because histone H2B is always associated with all parts of the genome
In ealry development of an embryo...
cells go through such rapid cell division that there is no time to do transcription so the first thing that you get from mothers is maternal messenger RNA as well as some mitochondria and a few proteins
mothers in all species load eggs (oocytes)...
with huge amounts of maternal mRNAs but most of that RNA is not active for trasnlation, once fertilization happens, then trasnaltion increases 100x and maternal mRNAs lengthen tehir polyA tail
the lengthening of maternal mRNAs polyA tail is a cause of...
translation increasing
poly A tail assay procedure (how to measure length of poly A tail)
Warning: You can't run a Northern Blot to measure the change in length of the polyA tail of a gene because the changes in te PolyAtail are very short compared to the length of the full gene. Also the length of the polyA tail can vary a lot even withing a single gene so we need a mroe precise way to measure
Steps:
1. Typically primers are 20 bases long but if a polyAtail is 100-200 bases, the primer could sit anywhere along the tail which goves us a whole range of PCR product, the short product will be favored and we will get an inaccurate measurement of the tail
2. Use Poly A polymerase to make the tail even longer and add G bases to the 3' end of the tail
4. We add a primer that is complementary to the G-A junction
5. The primer will be 5' CCCC-TTTT 3'
6. Use Reverse trasncriptase priming with the CT primer to move back towards the 5' end of the mRNA and form a DNA version of the gene mRNA
7. let the strands separate
8. Use a gene specific primer, TAQ polymerase, and dNTPS to amplify the DNA with TTTT and CCC
9. Heat that up to separate the original DNA from the PCR amplified one
10. Add primers gene specific primer (for original DNA) and the TC primer (for new DNA from PCR) to amplify again
11. To visualize add one radioactive nucleotide
12. Run on gel (DNA)
13. Depenidng on how long the poly A tail was to begin with is how long the PCR product will be
Using the Poly A tail assay to study bicoid mRNA during embryo development is the data RNA or DNA on the gel?
DNA the PCR amplified product is DNA because of the reverse transcriptase we use to turn mRNA into DNA
what is bicoid RNA?
it encodes a Trancription Factor and it is an example of maternal mRNA
Can you figure out how long the poly A tail is from looking at the gel?
No, because we don't know where the gene specific primer was. But, we could figure out how long the polyA tail was lengthened but not how long it is
If we wanted to know how long the polyA tail was we would have to knwo...
where on the map of teh gene the gene specific primer was. If you haad done this in teh lab you would know because ypu designed teh primer so you know where it sits on the gene
In the real world diagram of the bicoid mRNA polyA tail length we see a lot of smearing because..
there is variability between individual RNA molecules of the exact length of the poly A tail... the length is a range of distributions
How does mRNA polyA tail lengthening affect trasnaltion?
A mRNA starts getting translated:
1. The Poly A binding protein binds to the poly A tail (PABC)
2. the eIf4 complex binds to the 5' end and is stabilized by the Poly A binding protein
3. the small subunit and the large subunit come along
If you have a very short poly A tail...
you have very liitle poly A binding protein (PABC)
Translationally dormant to translatioanlly active process
1. CPEB (cytoplasmic polyadenylation element binding protein) binds to CPE (cytoplasmic polyadenylation element UUUUAU)
2. CPEB is phosphorylated which makes it lose the Maskin protein that was bound to it
3. Now CPEB recruits the Poly A polymerase (PAP) to lengthen the Poly A tail
5. PABC binds to the Poly A tail
6. eIF4 can recruit the other complexes
4. Now the RNA is trasnaltionally active
5. The lengthening of the poly A tail allows all teh initiation machinery to bind and transalte the RNA
You can have a whole set of RNA that are bound by CPEB that can...
switch from being translatioanlly dormant to active
What happens to bicoid mRNA after 4.5 hrs?
the poly A tail is getting shorter because the maternal mRNA are getting degraded. The embryo degrades maternal mRNA because at some point cell division slows down and cells want to make their own transcripts/ want to be different. Cells now use zygotic mrNA (mRNA transcribed in embryo) and not maternal mRNA
How would you design a system that degrades maternal mRNA but not degrade zygotic mRNAs?
put a specific signal in the 3'UTR to tag the maternal mRNA but not the zygotic mRNA
What are microRNAs?
short RNAs/ 21-23 nuclotide RNA molecules that base pair with mRNA
what is the experiment for degradation of maternal RNA?
is mature miR-430 involved in causing the degradation of maternal RNA?
If we do not activate zygotic trasncription, what would happen to the maternal RNA? - maybe degradation is dramatically slowed
miR-430 family
miR-430a, miR-430b, and miR-430c and thee are two large clusters on chromosome 4 one with 57 copies and another with 10 copies
Instead of mutating a large chunk of the genome...
they mutated part of the enzymatic machinery that creates mature miR
Seed pairing microRNAs:
have a seed that they use to base pair with target mRNA. they base pair from bases 2-8 and then after it doesn't have to base pair perfectly (there can be bubbles). BAse pairing at 5' end of microRNA is what is importnat for seed pairing
it leads to:
- mRNA deadenylation (removal of poly A tail after this removal mRNA can be degraded by an exonuclease)
- less translation/ less stable mRNA
Extensive pairing microRNA
- complete pairing between microRNA and mRNA
- known as small interfering RNA
- the now double stranded RNA leads to cleavage and degradation
types of microRNA
seed pairing and extensive pairing (siRNA)
WHich type of miRNA has more specificity?
extensive pairing
WHich miRNA can target more genes?
seed pairing
WHich type of microRNA is better suited to clear a LARGE set of maternal mRNAs?
seed pairing
Experiment: What happens when we keep the miRNA levels low to the translatability of the maternal RNAs?
If you get rid of Dicer from the cells, you block the pathway and the cleavage of the pre-miRNA and you don't get any mature miRNA. In the experiment, they got rid of miRNA being produced in the zebra fish embryo. They used Dicer because it dices up the pre miRNA so when you don't have Dicer you don't dice the pre miRNA and you don't have mature miRNA that can then interact with the mRNA.
Steps of experiment: Does miR-430 prevent translation of maternal RNAs?
1. they took a translational reporter gene (GFP- 3'UTR' PolyA signal)
2. Inject the translational reporter gene into fertilized eggs with wildtype 3'UTR
3. In fertilized eggs with wildtype 3'UTR there is no GFP so there is no translation
4. In fertlized eggs without Dicer, you see GFP so there is translation
5. Therefore, the whole miRNA pathway is important for repressing translation of the GFP reporter gene
In the experiment, why did they have to inject the mRNA and not put it in a plasmid?
No trasncription. At early stages of embryo development there is very little transcription. So if you put it in a plasmid, it would need to be transcribed to be able to see the reporter. If you inject the mRNA it only has to get translated and not transcribed first
Sofia explanation:
In the experiment, we saw no GFP with dicer: if dicer is present, there will be mature miRNA which binds to mRNA which leads to less translation therefore no GFP
We also saw GFP with no dicer: no dicer, no mature miRNA, no mature miRNA, miRNA does not bind to mRNA, mRNA is translated normally
When the zygote begins transcription, it produces these microRNAs, which bind to the maternal mRNA 3′UTR and trigger degradation.
What are the controls for the miR-430 experiment to test if it prevents translation of maternal RNAs? Specifically to conclude that microRNAs are working via the 3'UTR to prevent translation of the reporter gene?
a mutant 3' UTR to see if microRNA can still work with a mutated 3'UTR and an injection control
Results of control with mutated 3'UTR in the reporter gene
with a mutated 3'UTR miR-430 cannit bind as its binding site is mutated
1. the one with the wildtype dicer showed GFP expression
2. the one with no dicer showed GFP expression
Results of injection control (dsRef control- Poly A signal)
you can see red in all situations (it is ineffective)
- you use this to make sure that everything goes in equally and make sure mRNA got injected into the egg
How can specific mRNA (maternal mRNAs be degraded)?
different regulatory regions in the 3'UTR such as miRNA binding sites that lead to its degradation
How does microRNA binding affect an mRNA?
extensive pairing: cleavage (AGO2 complex)
seed pairing: the AGO1-4 complex recruits other molecules that ends up deadenylating at the 3' end of the mRNA and decapping the 5' end and then an exonuclease comes in and digests the mRNA. You can also get trasnaltional repression