Cancer Genetics
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
51 Terms
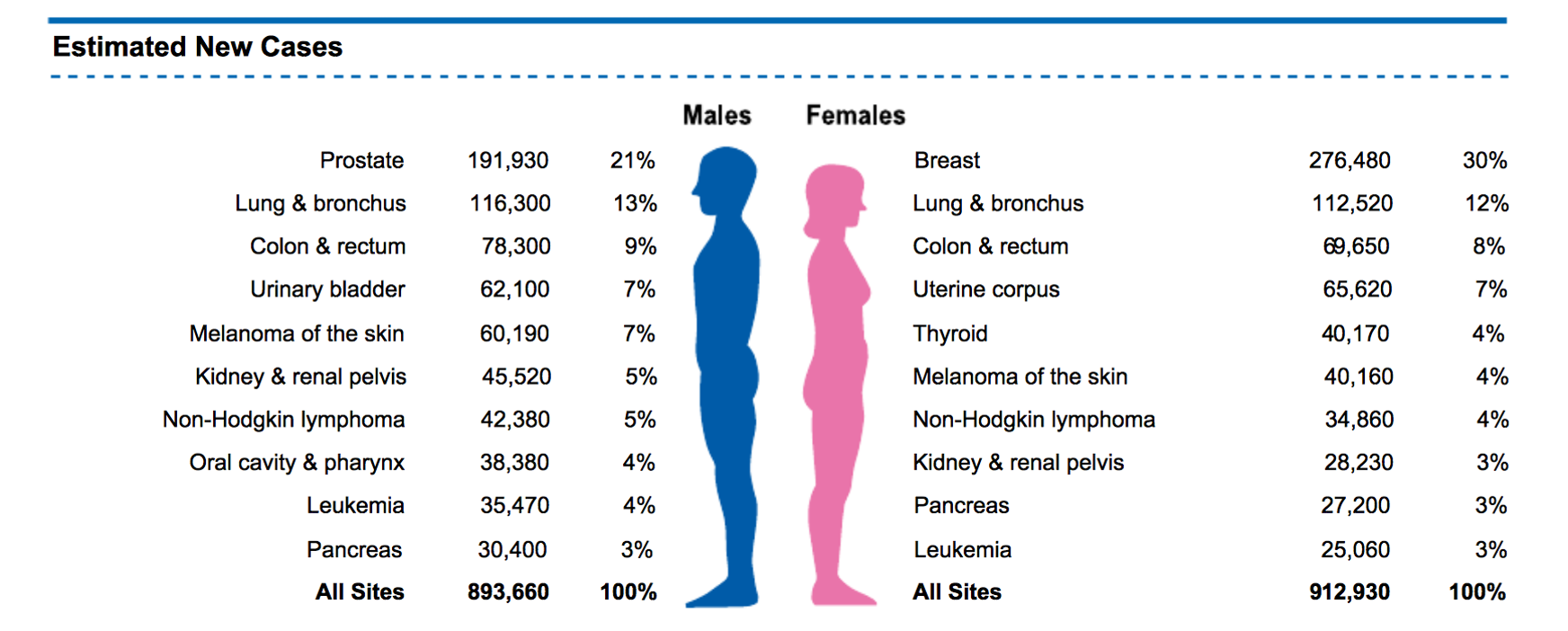
Cancer Incidence in 2020
1 in 3 Humans will develop cancer
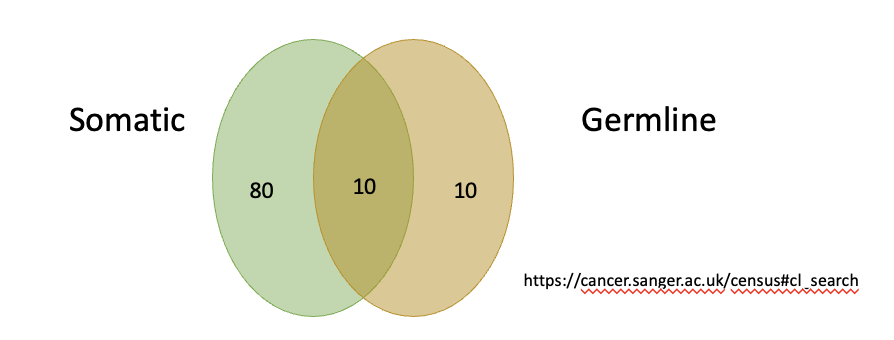
Gene Implicated Cancer
are specific mutations or alterations in DNA that contribute to the development of cancer.
>1% of all human genes are implicated via mutation in cancer.
Tp53 mutated in 50% of all cancers
Senescence
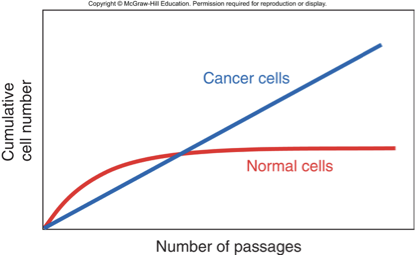
Cells cannot divide anymore
Fetal cells reach senescence after 40-60 divisions in culture
Senescence in Cancer Cells
Can bypass senescence by suppressing tumor suppressors
E.g., p53 and RB1 retinoblastoma protein
Cells reach crisis state due to short telomeres
Chr. ends seen as DSB, fusion, followed by more fusion and breakage, genome instability
Escape: reactivation of telomerase
Cancer Cells Acquire Immortality
•Cancer cells divide indefinitely
•Expression of telomerase in cancer cells maintains the length of chromosome ends
Higher Mutation Rate of Cancer Cells
Enzymatic systems that repair DNA damage or mistakes during replication are often defective.
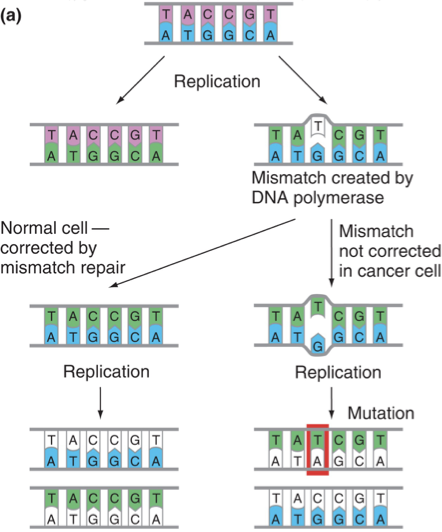
Genomic Instability
A hallmark of cancer characterized by a high frequency of mutations within the genome, often resulting from defects in DNA repair mechanisms.
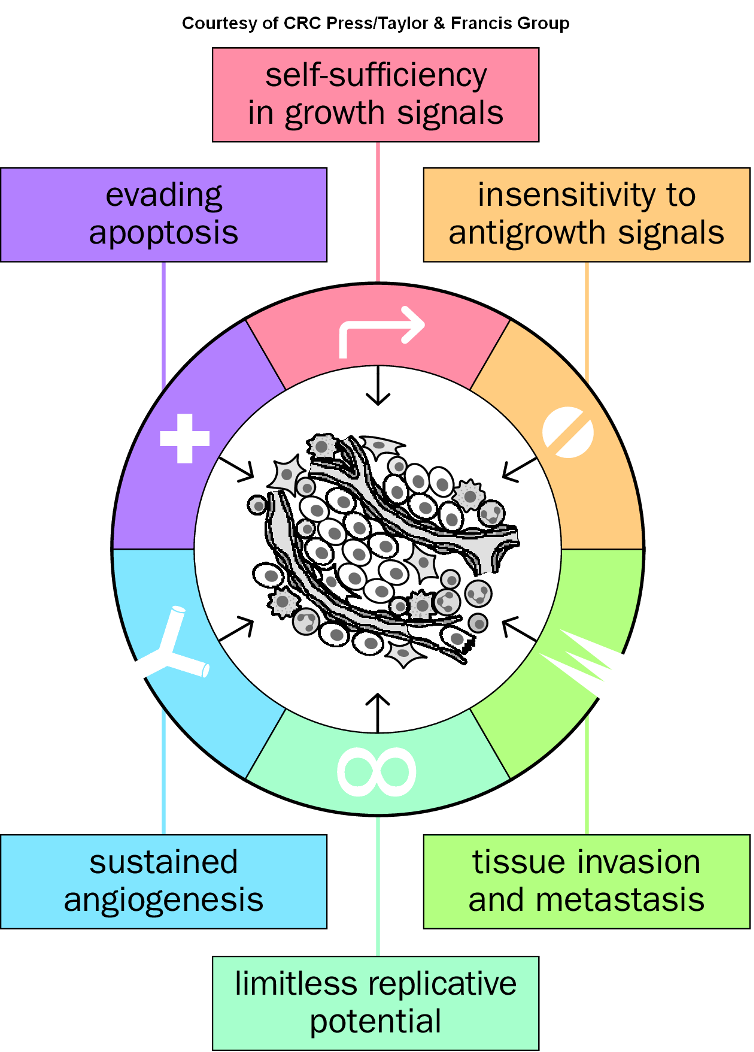
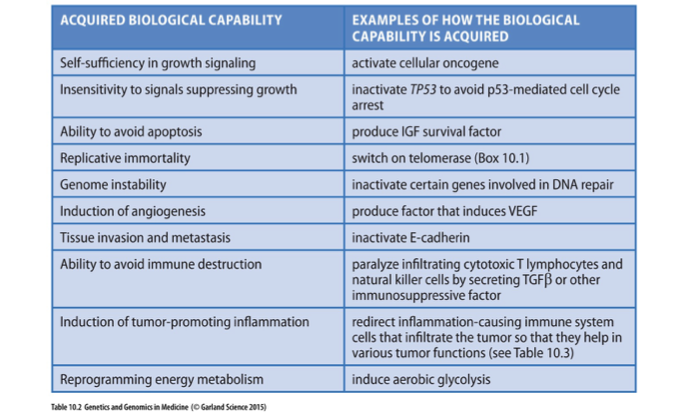
Hallmarks of Cancer (10 acquired properties)
Characteristics that define cancer progression, including uncontrolled cell growth, evasion of apoptosis, and sustained angiogenesis.
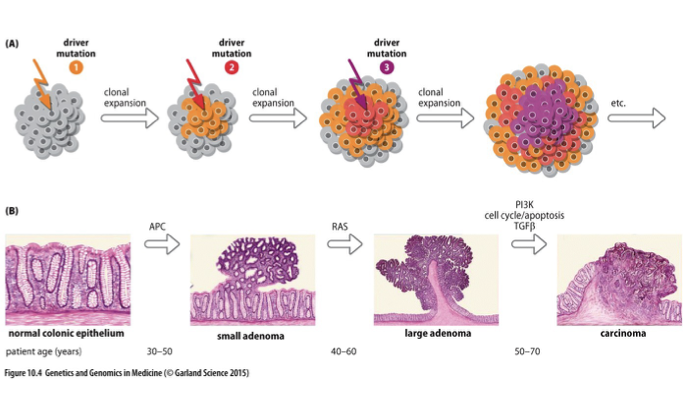
Multistage Evolution of Cancer
The process by which cancer develops through multiple genetic alterations over time, leading to progressively more aggressive tumor phenotypes.
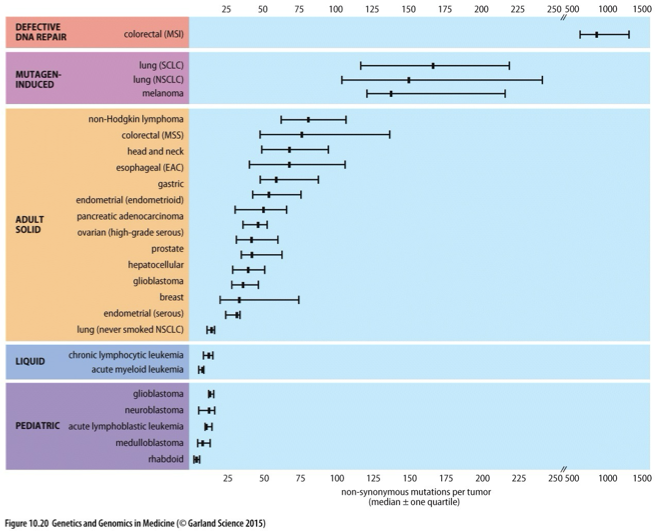
Non-synonymous Mutation Rate in Different Tumors
A measure of the frequency of mutations that alter amino acid sequences within proteins across various types of tumors, impacting cancer biology and treatment outcomes.
Clonal and Intercellular Heterogeneity
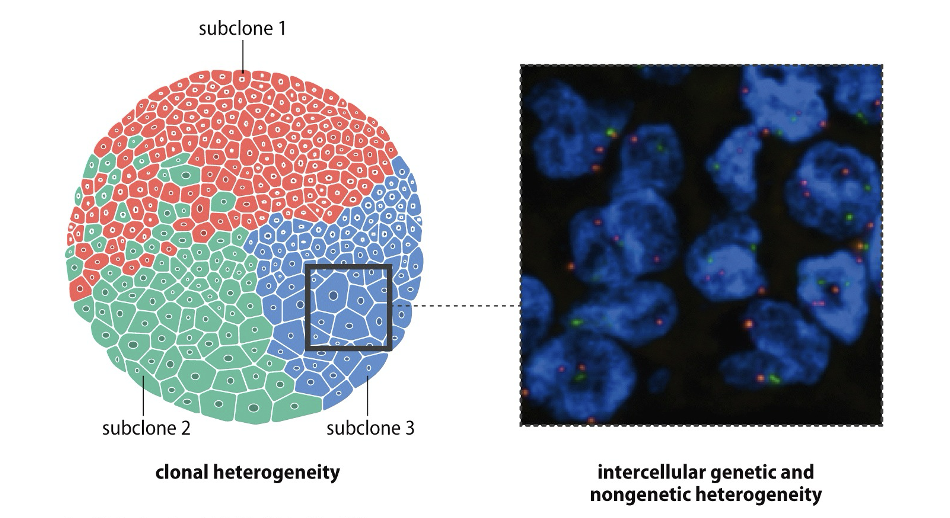
The presence of genetic diversity within a tumor and among its cells, leading to varied responses to treatment and differences in tumor behavior.
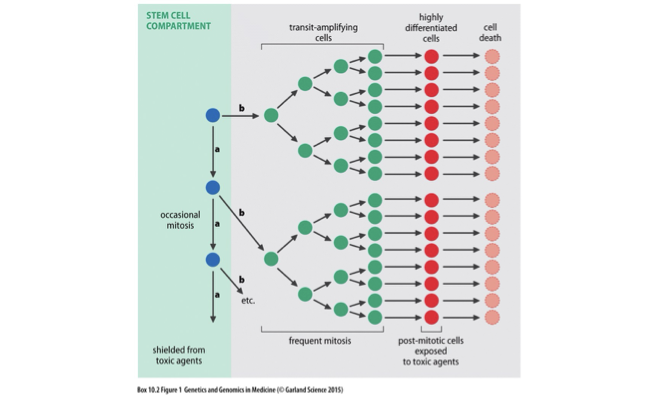
Cancer as Disease of STEM cells
Cancer originating from mutations in stem cells, leading to uncontrolled growth and differentiation, resulting in tumor formation.
Properties of Cancer Cells
•Readjust their metabolism
•Increased flux through pentose phosphate pathway
•Elevated rates of lipid biosynthesis
•Utilization of high rates of glucose
•Derive energy from glycolysis
•Epigenetic programming
•Cells may become less differentiated
•Driver and passenger mutations
•Development of counter-attacking measures to escape destruction by the immune system
•Recruitment of normal cells for support
•Metastasis
•Angiogenesis
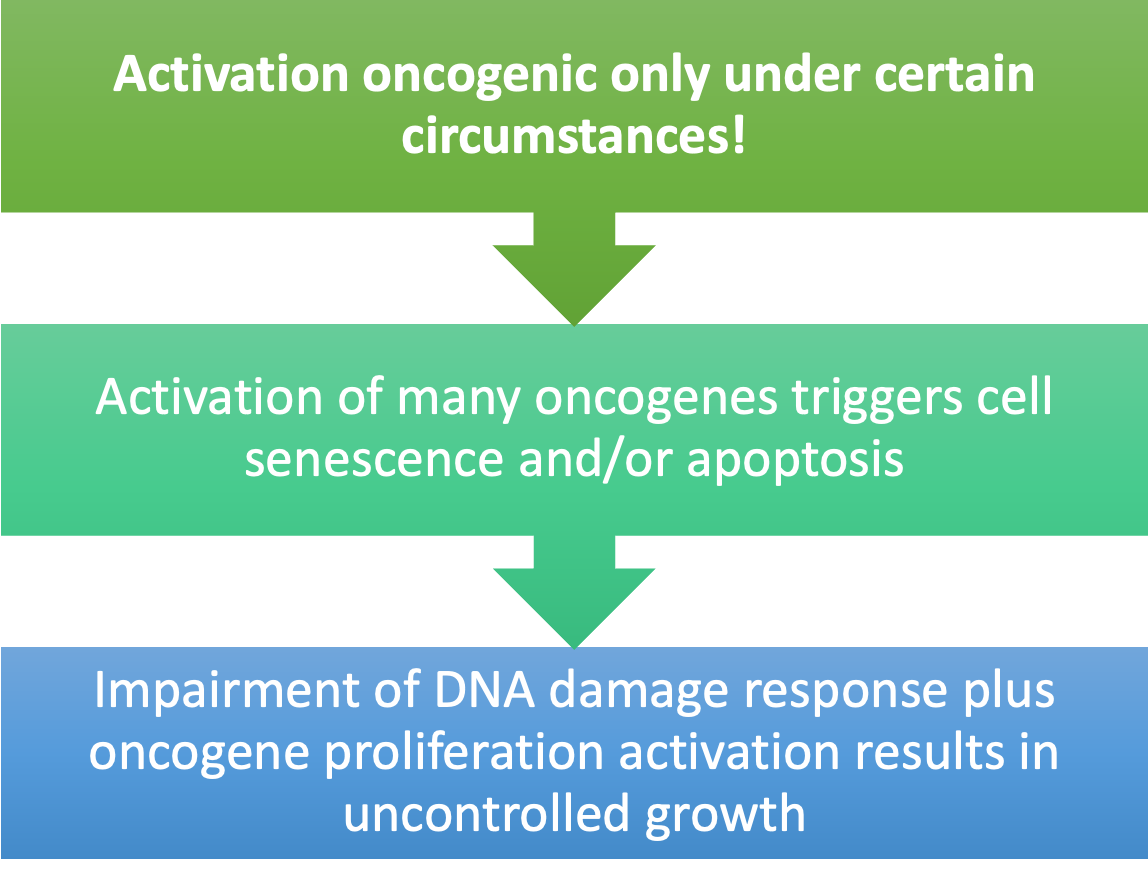
Oncogenes
•Dominantly acting cancer-susceptibility genes
•Often have role in growth signaling pathways to promote cell proliferation or inhibit apoptosis (proto-oncogenes)
•Growth factor
•Receptor
•Transcription factor
•Regulatory protein
•Suppression of apoptosis
•Activating mutation in single allele (gain-of-function)
Retrovirus-transforming Retrovirus Structure
A type of virus that can insert its genetic material into a host cell's DNA, often leading to the activation of oncogenes and promoting cancer development.
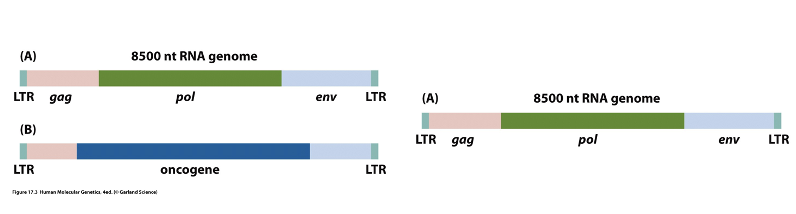
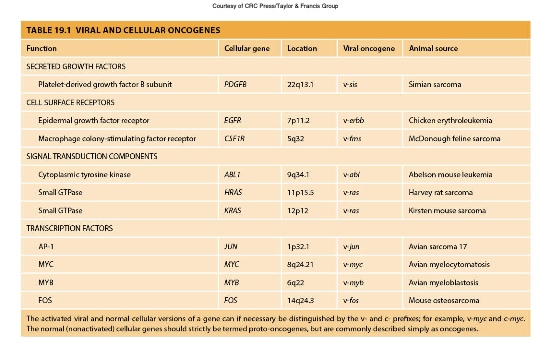
Viral and Cellular Oncogenes
Oncogenes derived from viruses or mutated versions of normal genes that promote cell growth and proliferation, contributing to cancer.
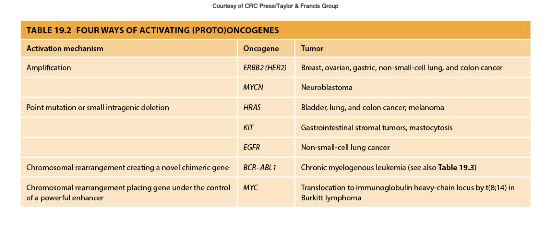
From Proto-oncogene to Oncogene or Oncogene Activation
Activation through gene amplification
Large number of copies
Result of complex rearrangements that bring together sequences from several different chromosomes
Extrachromosomal: double minutes
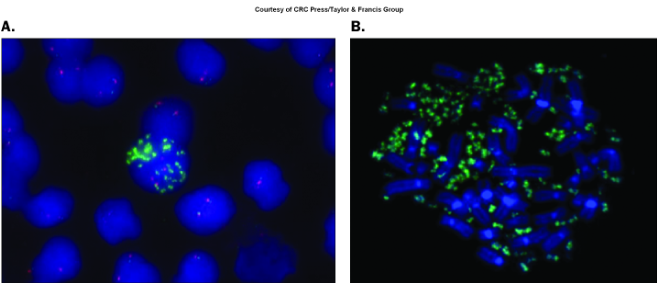
Tiny paired acentric chromatin bodies
Separated from chromosomes
Multiple copies of a small set of genes
Intrachromosomal: homogeneously staining regions
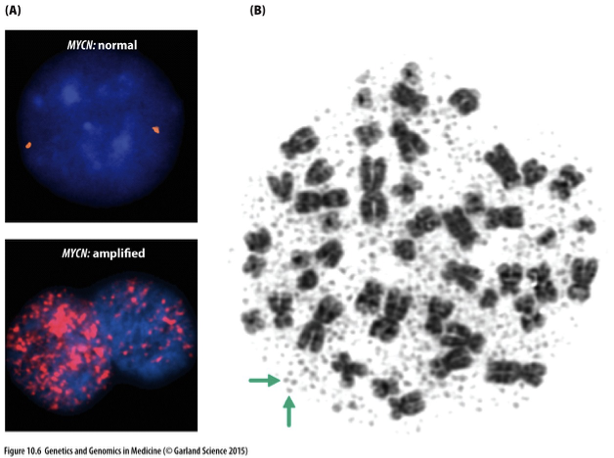
MYCN Oncogene in Neuroblastoma
A gene that, when amplified or mutated, plays a significant role in the development and progression of neuroblastoma, a childhood cancer arising from nerve tissues.
From Proto-oncogene to Oncogene/ Oncogene Activation
Activation through point mutation (in particular missense mutation)
Gain-of-function mutations
Some oncogenes are commonly mutated in cancer
Some mutations are recurrent only in certain cancers
Commonly recurrent mutations in certain regions of protein
Activation through specific changes
E.g., BRAF oncogene, which encodes tyrosine kinase
P.V600E accounts for 80% of all BRAF mutations
From Proto-oncogene to Oncogene
Epidermal growth factor receptor (EGFR or ERBB1)
Receptor kinase
Mutation in ATP-binding pocket common
Enhance signal
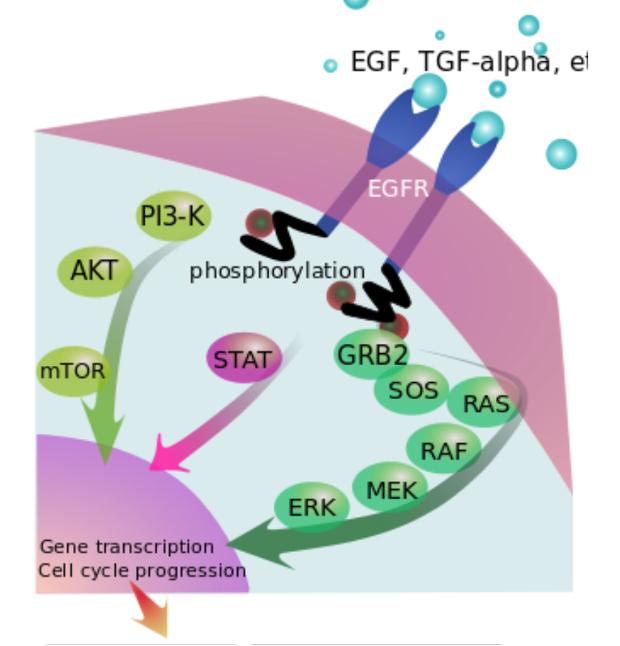
Ras (rat sarkoma) oncogene family
Human three genes (HRAS, KRAS,NRAS)
Work as GTPases and mediate growth signaling
Signaling hubs
1 in 6 human cancers has activating mutation in one of the RAS genes (most commonly KRAS)
>99% of mutations are in one of three codons:
12 [Gly], 13 [Gly], 61 [Gln]
![<p>Human three genes (HRAS, KRAS,NRAS)</p><p>Work as GTPases and mediate growth signaling</p><p>Signaling hubs</p><p>1 in 6 human cancers has activating mutation in one of the RAS genes (most commonly KRAS)</p><p>>99% of mutations are in one of three codons:</p><p> 12 [Gly], 13 [Gly], 61 [Gln]</p>](https://knowt-user-attachments.s3.amazonaws.com/2f52b4db-5f49-47f1-8be6-e4572d5388d3.png)
Translocation-induced gene activation
•Common in cancer
•1) Formation of chimeric/fusion gene
•Expression of oncogene sequence
•Philadelphia chromosome
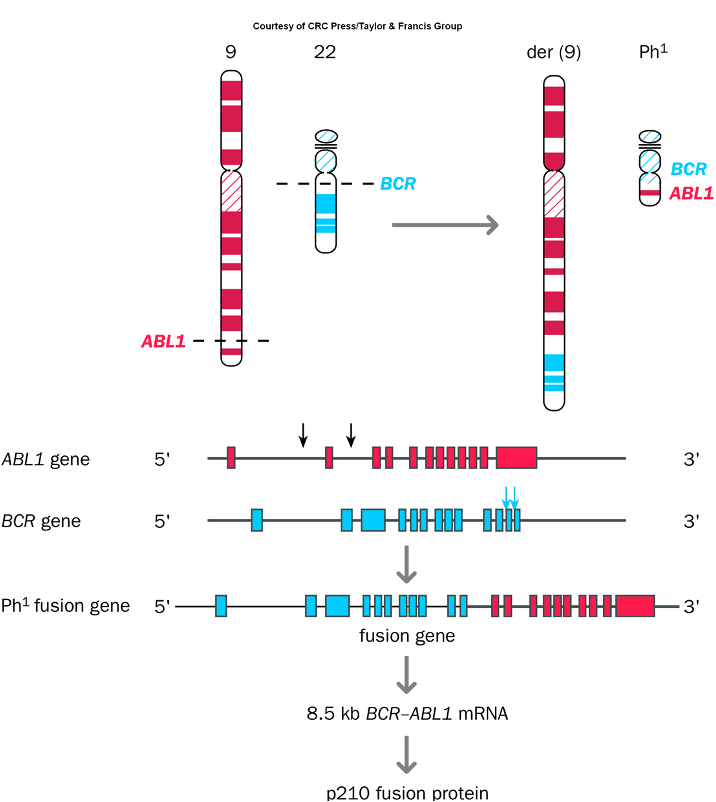
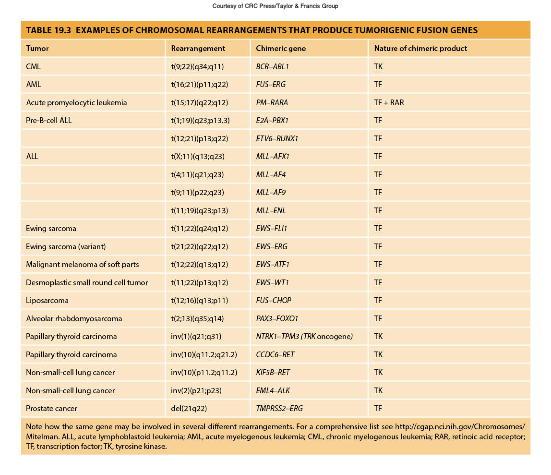
Chromosomal Rearrangements
Genetic alterations caused by breaks in chromosomes that lead to translocations, deletions, or inversions, often resulting in cancer by activating oncogenes or inactivating tumor suppressor genes.
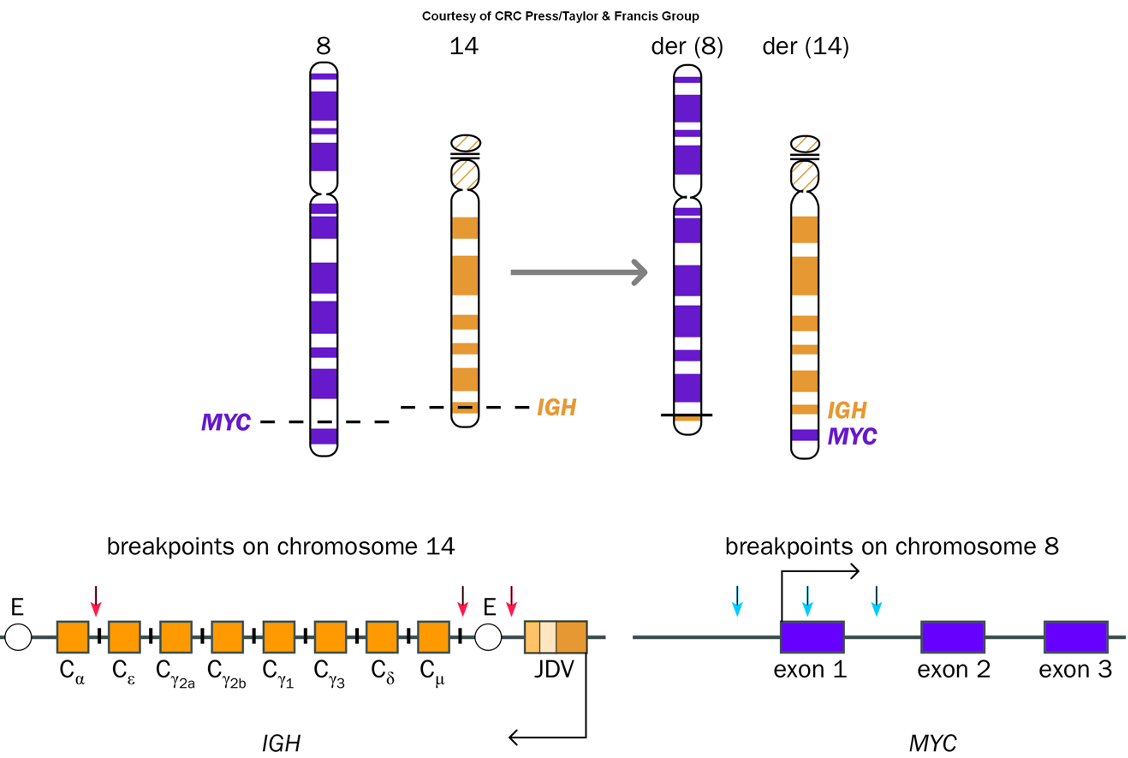
Enhancer capture
•Gene located in close proximity to regulatory sequence
•Tumors of B and T cells (breakpoints in immunoglobulin light or heavy chain (IGH), or T-cell receptor gene (TRA, TRB, TRD))
Activation of MYC Oncogene by Capture of B-cell-specific Enhancers
A process where specific enhancers from B-cells activate the MYC oncogene, often linked to chromosomal translocations in cancers such as Burkitt lymphoma.
Activation of Proto-Oncogenes
The process in which normal proto-oncogenes become oncogenes through mutations or chromosomal rearrangements, contributing to uncontrolled cell growth and cancer development.
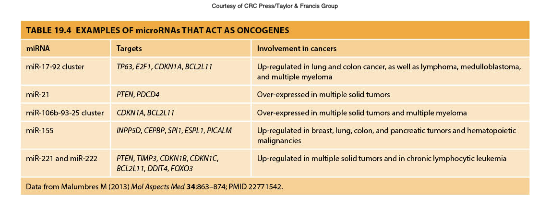
microRNAs as Oncogenes
•Aberrant miRNA expression very common
•One miRNA can target many mRNA species and single mRNA can be regulated by many miRNAs
•Difficult to identify specific miRNAs as key players in cancer
MicroRNAs (miRNAs) can function as oncogenes when their expression is dysregulated, leading to the modulation of multiple target mRNAs involved in cancer progression. These regulatory RNAs can influence various cellular processes, complicating the identification of specific miRNAs critical to cancer.
Oncogenes
Cell Proliferation Regulation Through Cancer Gene Classes
The mechanisms by which different classes of cancer-related genes, such as oncogenes and tumor suppressor genes, control the growth and division of cells, impacting tumor development and progression.
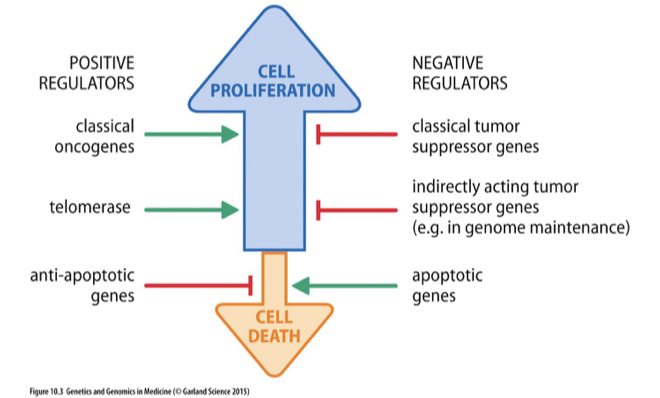
Tumor Suppressor Genes
•Restrain cell proliferation (opposite to oncogenes)
•Gatekeeper genes:
•Direct control through
•Regulation of cell cycle and induction of cell cycle arrest
•Role in upstream growth signaling pathways
•Promote apoptosis
•Caretaker genes:
•Help maintain integrity of the genome
•Landscaper genes:
•Control stromal environment in which the cells grow
•Contribute to cancer if gene is lost or inactivated
•Commonly recessive cancer-susceptibility genes
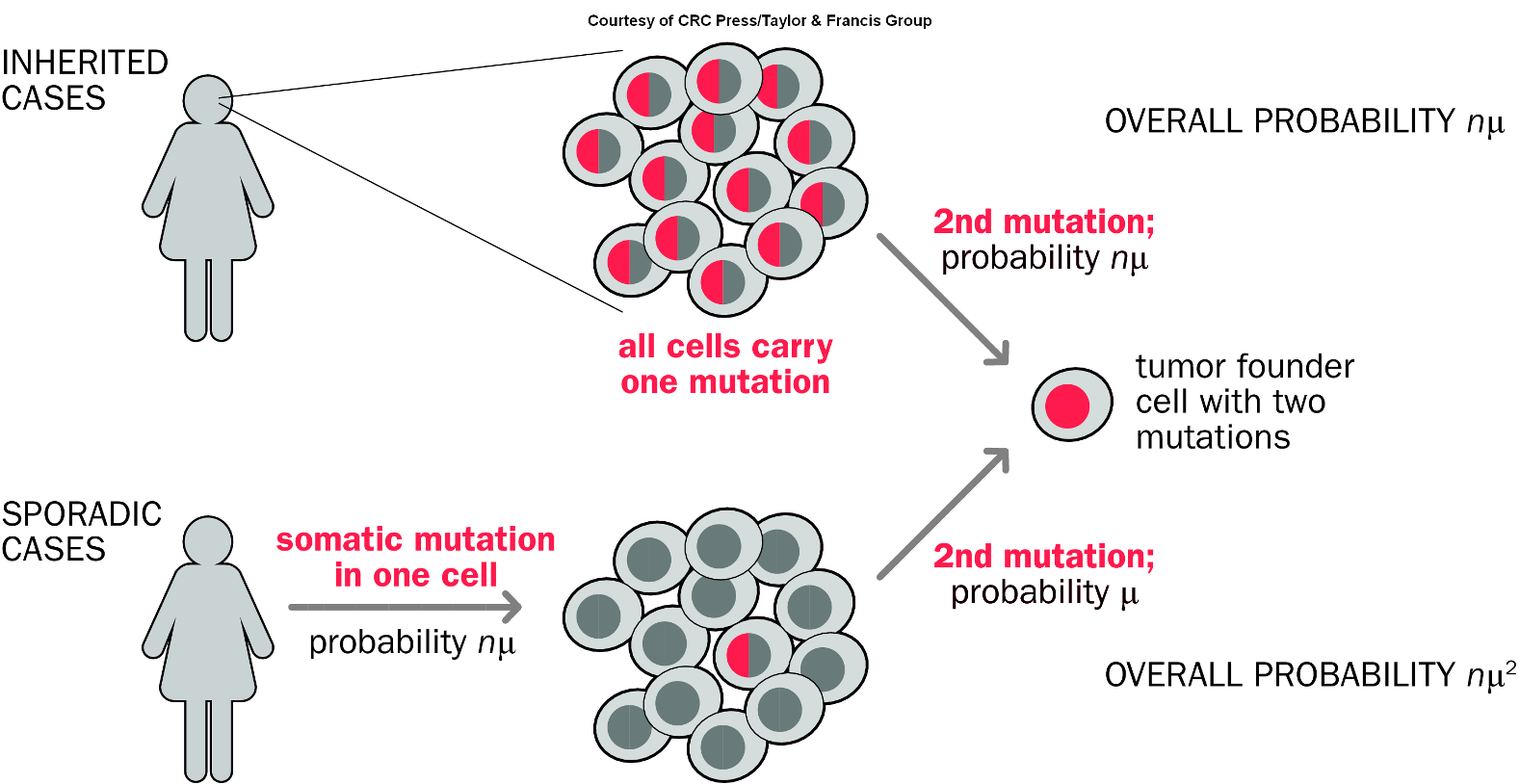
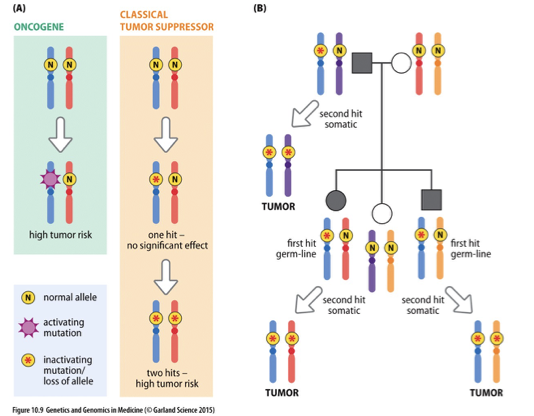
Knudson’s 2 Hit Hypothesis
A model that explains how two genetic mutations in tumor suppressor genes can lead to cancer, suggesting that both alleles of a gene must be inactivated to promote tumorigenesis. It posits that one inherited mutation (the first hit) is not sufficient to cause cancer, as the second mutation (the second hit) must also occur in the other allele for cancer to develop.
Retinoblastoma
•Caused by mutations in RB gene
•Inheritance of one RB− copy predisposes to cancer of retina
•Often tumors of both eyes
•During proliferation of retinal cells, RB+ allele is lost or mutated
•Tumors develop as clone of RB−/RB− cells
a childhood eye cancer characterized by abnormal cell growth in the retina, typically associated with mutations in the RB1 gene.
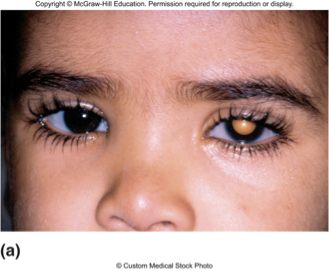
Confirmation of Knudson’s Two Hit Hypothesis
Mitotic recombination
Deletion
Loss of gene through mutation
Loss of chromosome and duplication
or loss of heterozygosity
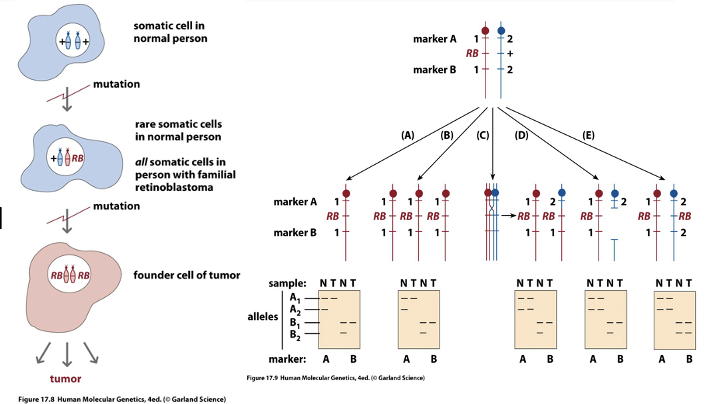
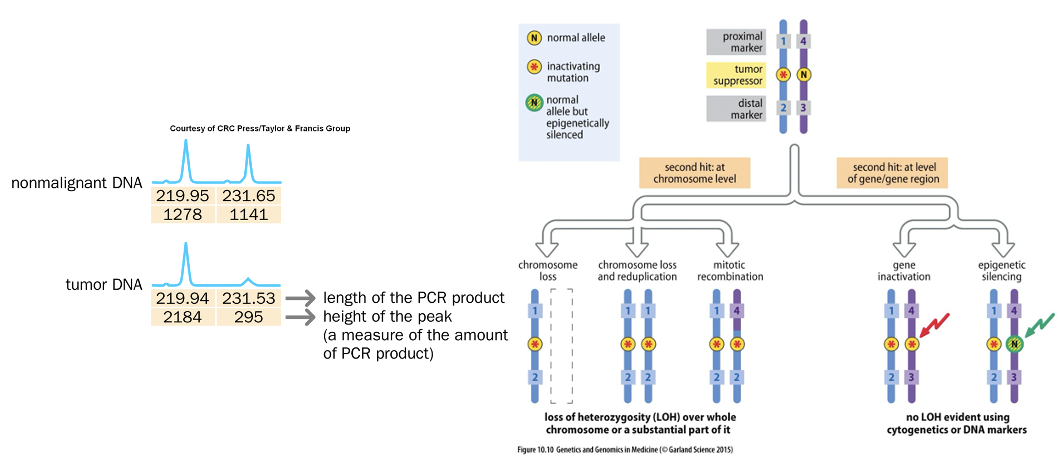
Second Hit at Tumor Suppressor Locus
refers to the additional genetic alteration that occurs after the first mutation in a tumor suppressor gene, essential for tumor development.
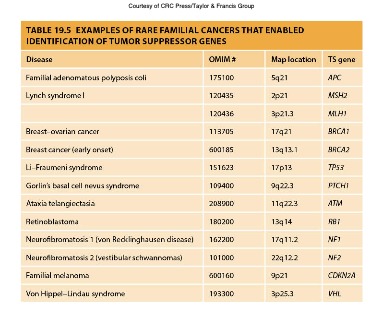
Familial Cancers and Tumor Suppressor Genes
are types of cancers that occur due to hereditary mutations in tumor suppressor genes, increasing the risk of developing specific cancers in affected families.
Tumor Suppressor Gene Silencing
•Through mutation
•Often silenced epigenetically through methylation
or histone modification, reducing the expression of crucial genes that control cell growth and division.
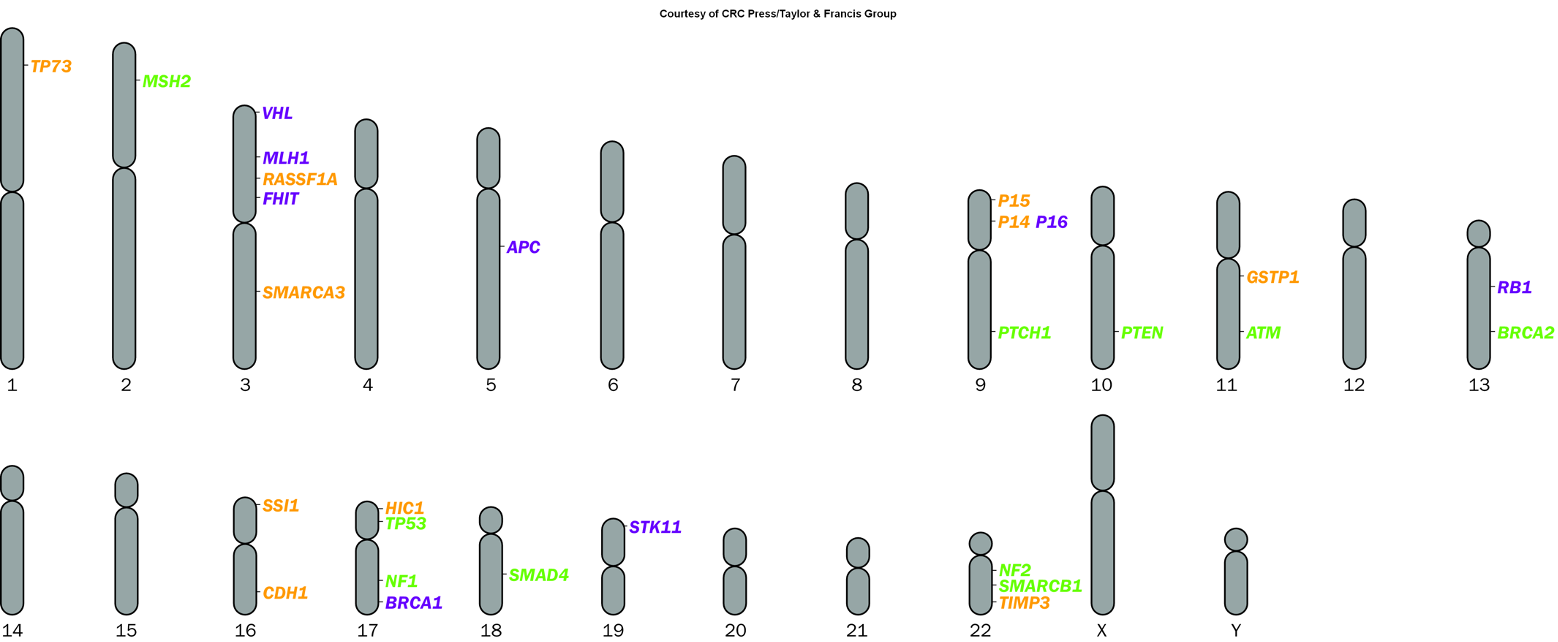
Oncogene vs. Classical Tumor Suppressor Gene
Oncogenes are genes that, when mutated or overexpressed, promote tumorigenesis, while classical tumor suppressor genes normally inhibit cell division and prevent tumor formation. The loss or mutation of these tumor suppressor genes leads to uncontrolled cell growth.
Oncogene or Tumor Suppressor Gene?
Oncogene
microRNA as Tumor Suppressor Genes
More common than the involvement of miRNA as oncogenes
and play a crucial role in regulating gene expression by targeting mRNAs for degradation or repression, thus preventing excessive cell proliferation.
2 Hit Hypothesis
Not true for all cancers
Some familial cancers have different muta-tions than majority of sporadic cancers (e.g., breast cancer)
Haploinsufficiency
Gain-of-function mutation
Partial loss of tumor suppressors can make considerable contribution to tumorigenesis
Loss of Cell Cycle Control
Many types of cancer result from faulty checkpoints
Timing, rate, and number of cell divisions depend on:
Protein growth factors
Signaling molecules from outside the cell
Transcription factors within
G1-S Cell Cycle Checkpoint
Regulates entry from the G1 phase to the S phase in the cell cycle, ensuring that the cell is ready for DNA replication and that any damage is repaired before proceeding.
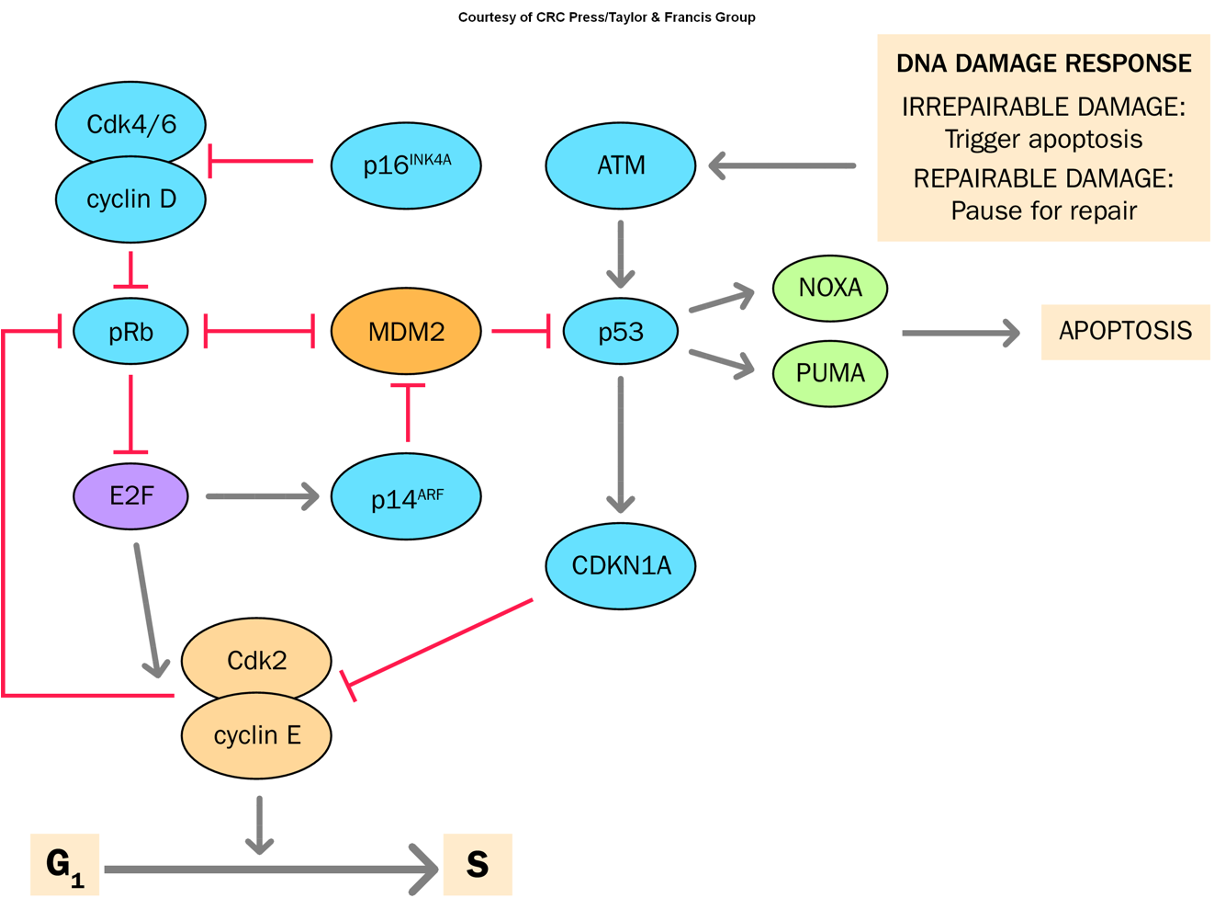
CDKN2A
A tumor suppressor gene that regulates the cell cycle by inhibiting cyclin-dependent kinases, playing a crucial role in preventing uncontrolled cell proliferation.
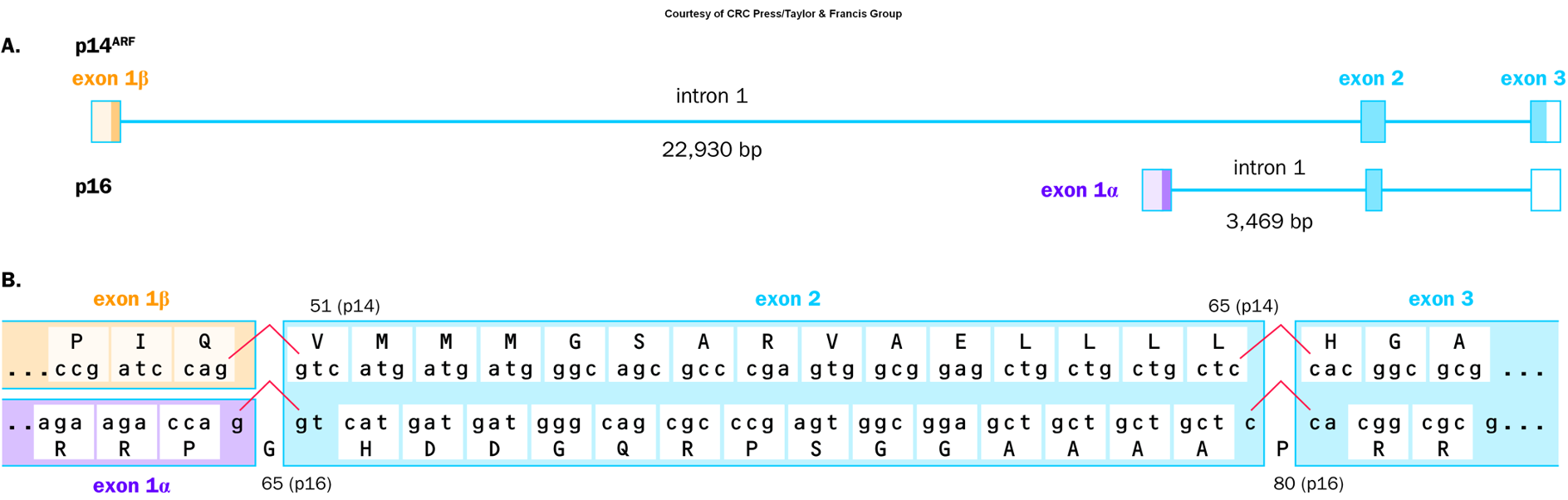
Mutations in this gene are associated with various cancers, including melanoma and pancreatic cancer.

p53
Guardian of the genome
Determines if a cell repairs DNA replication/damage errors or dies by apoptosis
More than 50% of human cancers involve an abnormal p53 gene
Expressed in virtually all cells
Self-limiting regulation
Non-classical tumor suppressor gene
Li-Fraumeni syndrome
Family members have a very high risk of developing cancer
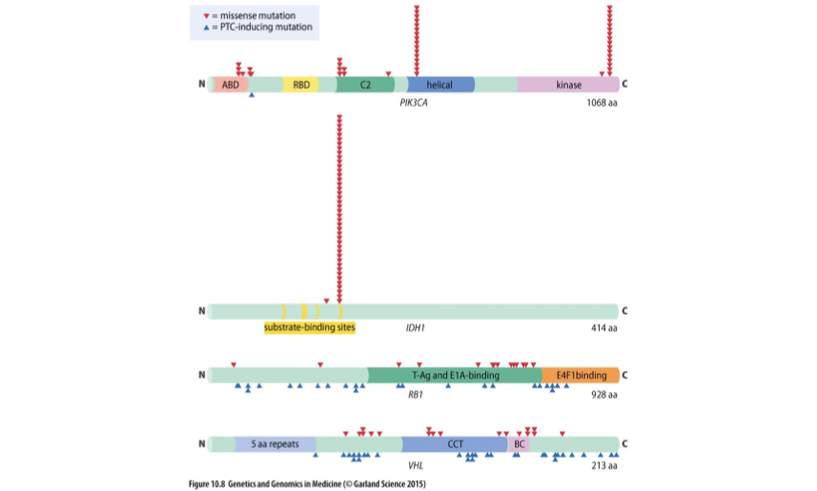
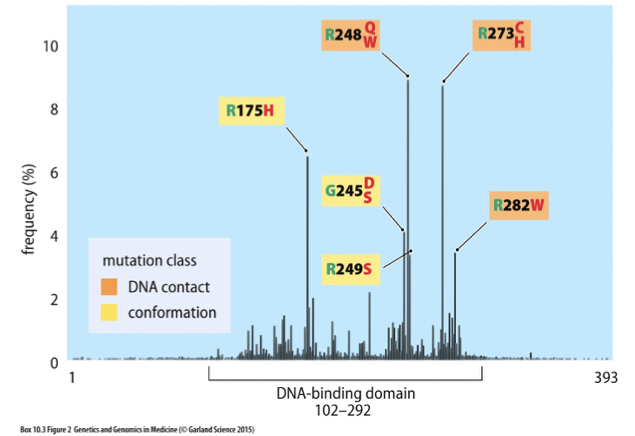
Tp53 Mutation Spectrum
Also known as p53, it is a tumor suppressor gene crucial for regulating the cell cycle, DNA repair, and apoptosis. Mutations in Tp53 are commonly associated with various cancers.
Li-Fraumeni Syndrome
A hereditary condition caused by mutations in the TP53 gene, leading to a significantly increased risk of developing multiple types of cancer at a young age.
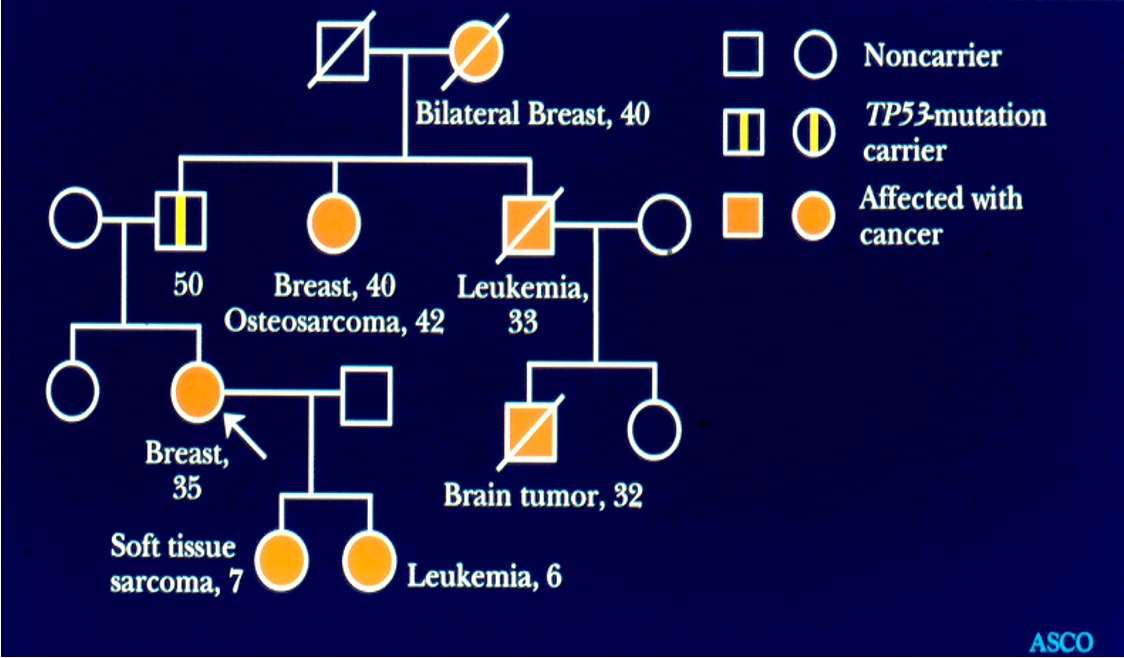
Li-Fraumeni Syndrome
Sarcoma (soft tissue and osteo)
Pediatric leukemias, adrenocortical carcinoma
Breast cancer
Brain cancer
Cancer at an earlier age than expected
Diagnosis of more than one cancer over lifetime
Autosomal dominant
80% have mutation in TP53
Peutz Jeghers Syndrome
Benign polyps called hamartomas on mucous lining of gastro-intestinal system
Dark blue to dark brown skin freckling (melanocytic macules) around the mouth, eyes, nostrils, fingers, oral mucosa and anus (perianal)
Highly increased (up to 93%) risk of developing gastrointestinal and other cancers including breast, cervical, uterine, pancreas, testes, and lung
Most patients develop cancer by fifth decade
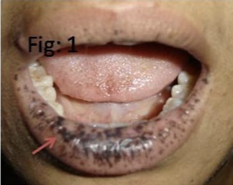
Disease Jegher Syndrome: Mode of Transmission
Autosomal dominant
Caused by mutations in STK11/LKB1 gene (80-94%)
>200 disease-causing mutations reported
Penetrance is thought to be 100%
Prevalence 1/50,000-1/200,000
Women at higher risk of developing cancers compared to men
PJS increases likelihood of developing breast, ovarian, cervical, and uterine cancer
Familial Adenomatous Polyposis (FAP)
Multiple colonic polyps, 100% risk colon cancer
Pancreas, thyroid, brain, hepatoblastoma
Autosomal dominant
Caused by mutations in the APC gene
Congenital hypertrophy of the retinal pigment epithelium (CHRPE)
Gardner syndrome (osteomas)
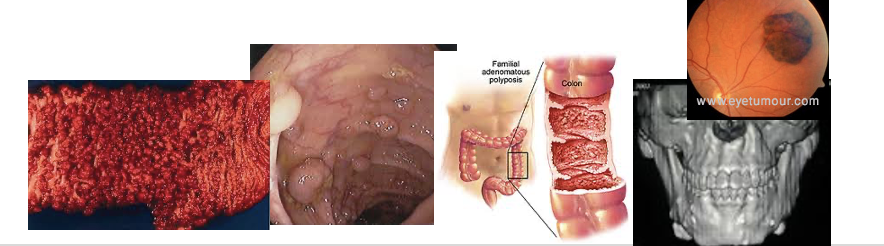
FAP in Family But No Mutation Screening/Identification
Anyone at risk for FAP but who has not had their diagnosis confirmed by genetic testing should be screened yearly, by sigmoidoscopy, from age 10 to 24.
At that point, screening should occur every two years until age 34.
Then every three years until age 44
And every three to five years thereafter