Biochem Sept. 22nd
1/30
Earn XP
Description and Tags
Topic 5
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
31 Terms
What are Enzymes?
Enzymes are biological catalysts that accelerate or regulate most chemical transformations within cells
They participate in chemical reactions but are not altered themselves
Most are proteins, though some are RNA molecules known as ribozymes
Are enzymes efficient?
Yes, they can turn reactions that might take years, into seconds or faster
Able to work under mild conditions
What is the Active Site?
The active site is a small specific surface where the substrate binds and the chemical reaction occurs
The substrate binds to the active site through non-covalent bonding interactions
E + S ⇌ ES —→ EP ⇌ E + P
S - substrate
P - product
Do enzymes change the equilibrium position?
No, they also don’t change the overall standard free energy change (ΔG) between reactants and products (only affect the rate)
What is the transition state?
An unstable, high-energy intermediate form that reactants must pass through to become products. It represents the point of highest free energy along the reaction pathway, where bonds are in the process of breaking and forming
What is the activation energy ( \Delta G^{\ddagger} )?
The free energy difference between the transition state and the reactants. A higher \Delta G^{\ddagger} means a slower reaction rate.
How do enzymes effect the activation energy?
Enzymes accelerate reactions by lowering the \Delta G^{\ddagger}, making it easier and faster for reactants to reach the transition state
What are the strategies enzymes use to lower the activation energy?
Proximity and Orientation
Enzymes bring the substrates together in the correct orientation to facilitate the reaction and stabilize the transition state
Charge Stabilization
Active site residues can use their own charges to influence the charge distribution of the substrate, making the bond breaking or formation easier
Stabilization of Transition State
Enzymes preferentially bind to and stabilize the transition state, lowering its free energy and thus reducing the overall \Delta G^{\ddagger}
What are transition state molecules?
Molecules that resemble the transition state but cannot be converted into product. They bind to the enzyme with very high affinity, often more tightly than the substrate itself, making them potent enzyme inhibitors.
What are Cofactors?
Non-protein partners required by some enzymes for function
Metal ions
Coenzymes
Co-substrates: coenzymes that bind transiently to the enzyme, often undergoing modification (oxidation/reduction) and then dissociating
Prosthetic Groups: Coenzymes that are permanently (tightly and stably) bound to the enzyme, similar to heme in myoglobin/hemoglobin
Describe Chymotripsin
Type: A member of the serine protease family, characterized by a critical serine residue in its active site
Source and Function: Secreted by the pancreas into the intestine to break down dietary proteins through hydrolysis of peptide bonds
Hydrolysis is the addition of a water molecule to break a peptide bond
What is the Catalytic Triad in Chymotripsin?
Serine 195
The nucleophile that attacks the carbonyl carbon of the peptide bond (attacker)
Histidine 57
Acts as a general acid-base catalyst, abstracting a proton from Ser195 to increase its nucleophilicity, and later donating a proton to stabilize the leaving group (amine) (Takes and gives protons)
Aspartic Acid 102
Stabilizes the positively charged His57 through a strong hydrogen bond, increasing its basicity and ability to abstract protons (supports/shapes His)
What is the 1st step in the mech. of chymotrypsin?
The substrate enters the active site of the enzyme.
His 57 steals a proton (H⁺) from Ser 195, making Ser’s oxygen highly reactive.
Now Ser 195 attacks the carbonyl carbon (C=O) of the peptide bond in the substrate.
This creates a tetrahedral intermediate (a temporary, unstable shape).
✅ Goal: Activate Ser so it can attack the peptide bond and form the first temporary intermediate.
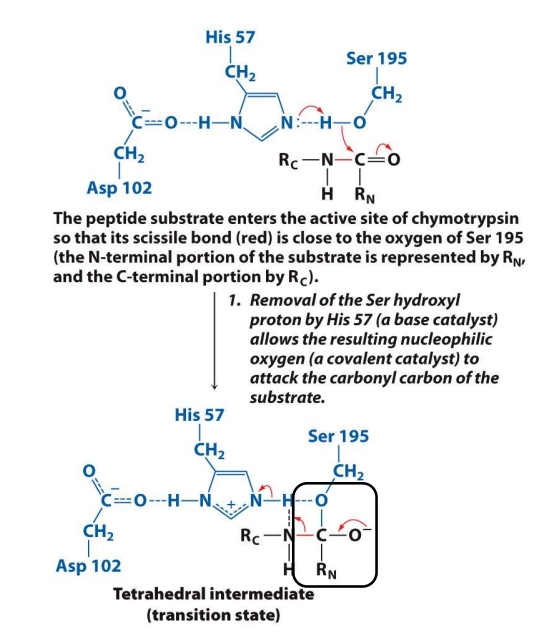
What is the 2nd step in the mech. of chymotrypsin?
The tetrahedral intermediate is unstable.
His 57 now gives a proton (acts acidic) to the nitrogen of the peptide bond.
This helps break the peptide bond, separating the substrate.
The C-terminal fragment (RC) leaves.
What remains is still attached to Ser — this is called the acyl-enzyme intermediate.
✅ Goal: Break the original peptide bond and release the first product (C-terminal piece).
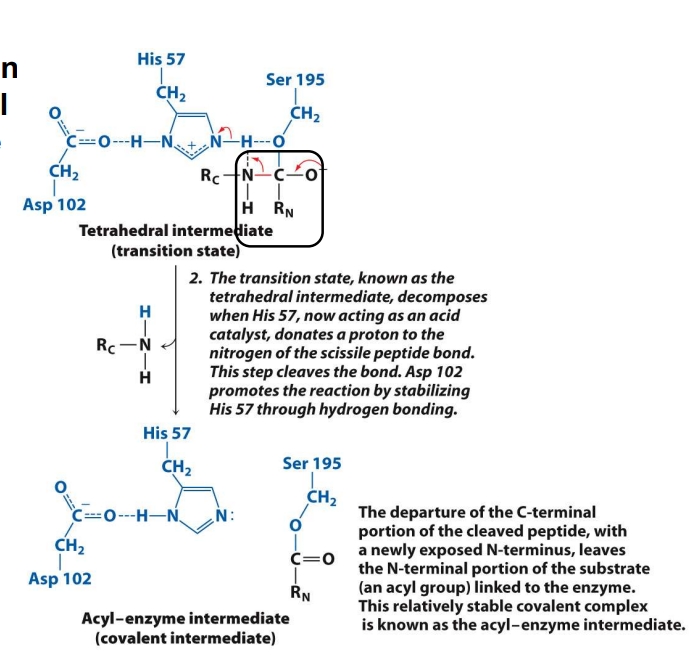
What is the 3rd step in the mech. of chymotrypsin?
A water molecule (H₂O) enters.
His 57 steals a proton (H⁺) from water, making water reactive (turns into OH⁻).
The reactive OH⁻ attacks the carbonyl carbon of the acyl-enzyme intermediate (just like Ser did earlier).
✅ Goal: Use water to begin breaking the link between the enzyme and the remaining part of the substrate.
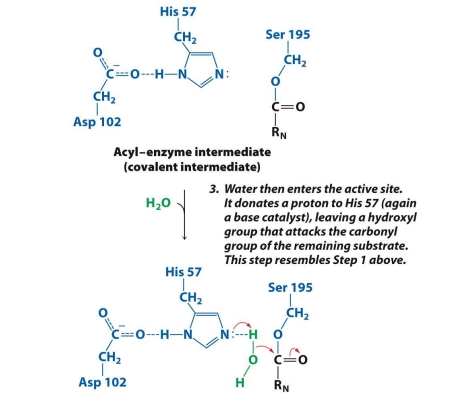
What is the 4th step in the mech. of chymotrypsin?
After water attacks, another tetrahedral intermediate is formed.
His 57 donates a proton back to Ser 195’s oxygen.
This helps the intermediate collapse.
✅ Goal: Collapse the intermediate so the second fragment can be released.
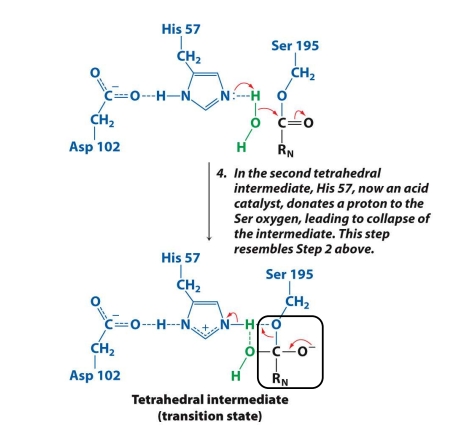
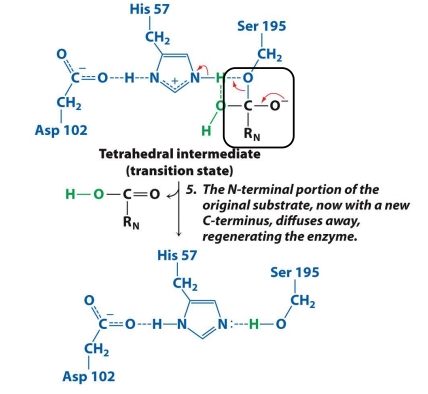
What is the 5th step in the mech. of chymotrypsin?
The N-terminal fragment (RN) is released (now with a new C-terminus).
Ser 195 is no longer attached to anything.
The enzyme returns to its original state, ready to work again.
✅ Goal: Release the second product and reset the enzyme.
What are the two phases of the mech. of chymotrypsin?
Phase 1: Enzyme attacks substrate (Uses serine to attack substrate)
This phase ends with the 1st product released (C-terminal end)
Phase 2: Water attacks enzyme-substrate complex to free enzyme again (like a reverse version of phase 1 where water is used instead of Serine)
This phase ends with the second product released (N-terminal end) and the enzyme regenerated
Whats the oxyanion hole and what does it do?
The oxyanion hole is a small pocket in the enzyme’s active site that stabilizes a negatively charged oxygen (an "oxyanion") that forms during the reaction
During the formation of the tetrahedral intermediates (in both Step 1 and Step 4), the carbonyl carbon of the substrate is attacked, and the carbonyl oxygen becomes negatively charged (O⁻)
The oxyanion hole stabilizes the negative charge by forming hydrogen bonds with the negatively charged oxygen. This lowers the energy of the intermediate → making the reaction faster and easier.
What are serines different proteases?
chymotrypsin
trypsin
elastase
They all use the same catalytic triad (Ser-His-Asp) but cut different kinds of peptide bonds because of their different specificity pockets
Whats a specificity pocket?
It’s a special pocket near the active site of the enzyme that recognizes and binds specific types of amino acid side chains on the substrate. This determines where the enzyme will cut.
What kind of amino acids does chymotrypsin like?
Large, hydrophobic/aromatic amino acids (e.g. Phe, Tyr, Trp)
because its pocket is deep and hydrophobic
What kind of amino acids does Trypsin like?
Positively charged amino acids (e.g. Lys, Arg)
Because it has a negatively charged residue (Asp) at the base of the pocket
What kind of amino acids does Elastase like?
Small amino acids (e.g. Ala, Gly)
Because pocket is shallow and blocked by bulky residues
What are zymogens?
Inactive precursors of proteases (like chymotrypsin and trypsin)
Are first made as zymogens so that they don’t start digesting proteins too early
Ex. Trypsinogen is the inactive form of trypsin
it becomes active trypsin only when an enzyme called enteropeptidase cuts it in the small intestine
This then activates chymotrypsinogen (partially active chymotrypsin), which then fully activates chymotrypsin (with small multiple cuts)
These cuts cause slight shape (conformation) changes that fully activate the enzyme.
What is the Michaelis-Menten equation?
v0 is the initial velocity (before any product is produced)
Vmax is the maximal velocity (when E is entirely in ES form)
[S] is concentration of substrate
When [S] is low, the reaction rate is low because the enzyme is not saturated with substrate and vice versa
KM is the Michaelis constant
A smaller KM indicates a tighter binding or higher affinity of the enzyme for its substrate
![<ul><li><p><strong>v0</strong> is the initial velocity (before any product is produced)</p></li><li><p><strong>Vmax</strong> is the maximal velocity (when E is entirely in ES form) </p></li><li><p><strong>[S]</strong> is concentration of substrate </p><ul><li><p>When [S] is low, the reaction rate is low because the enzyme is not saturated with substrate and vice versa</p></li></ul></li><li><p><strong>KM</strong> is the Michaelis constant</p><ul><li><p>A smaller KM indicates a tighter binding or higher affinity of the enzyme for its substrate</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/125bc7aa-2b3a-4b03-9740-d41ffff51632.jpg)
What are enzyme inhibitors?
Molecules that decrease the activity of enzymes by binding to them, preventing substrate interaction
There’s reversible inhibition or irreversible inhibition
Whats the mech. of enzyme inhibitors?
Enzyme inhibitors can work through various mechanisms, such as competitive inhibition, where the inhibitor competes with the substrate for the active site, or non-competitive inhibition, where the inhibitor binds to an allosteric site and alters the enzyme's activity without competing for the active site
Increases the KM and therefore, needs a higher concentration of substrate to reach the Vmax
The Vmax stays unchanged
What are allosteric regulators?
Molecules that bind to an enzyme at a site other than the active site, altering its activity
Allosteric regulators can either enhance or inhibit enzyme function, influencing metabolic pathways
Ex. Hemoglobin is an allosteric protein, where BPG binding at an allosteric site lowers its affinity for oxygen
Whats another example of an allosteric regulator other than BPG?
PFK: converts Fructose-6-phosphate → Fructose-1,6-bisphosphate
speeds up glycolysis early on
PEP (phosphoenolpyruvate) is formed later on in glycolysis and if too much PEP builds up, it binds to PFK, inhibiting its activity and slowing down glycolysis