05 - Organelles and ER: Quality Control and Secretory Pathway
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
74 Terms
Animal cells typically have the following membrane-contained organelles:
Nucleus
Endoplasmic Reticulum (ER)
Mitochondria
Golgi Apparatus
Peroxisomes
Lysosomes/Endosomes
Plant cells typically have the following membrane-contained organelles:
Nucleus
Endoplasmic Reticulum (ER)
Mitochondria
Golgi Apparatus
Peroxisome
Lysosome/Endosome
Vacuole
Chloroplasts
Cytoplasm
Contents of the whole cell that are within the plasma membrane but outside the nucleus.
Main site of protein synthesis/degradation
Cytosol
portion of cytoplasm that is outside membraneenclosed organelles
most of total cell volume
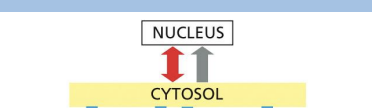
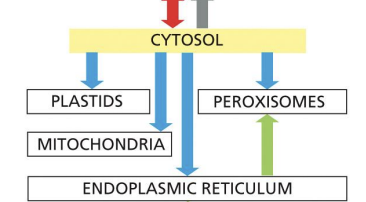
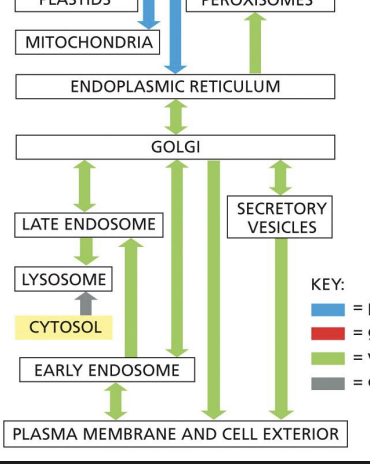
The intracellular compartments can be grouped into four distinct families:
1.Nucleus and cytosol – communicate via nuclear pore complexes
2.ER, Golgi, endosome, lysosome, peroxisomes and transport vesicles - function in endocytic and secretory pathways
3.Mitochondria
4.Chloroplasts (plants)
Mitochondria/Chloroplasts
generation of ATP.
Endoplasmic Reticulum (ER)
Most membrane protein and soluble organelle protein synthesis.
Golgi apparatus
Sorting of proteins and lipids from ER.
Nucleus
contains genome, DNA and RNA synthesis
Endosomes
Move most material between organelles and uptaken extracellular molecules
Lysosomes
Degradation of unneeded intracellular organelles and extracellular molecules that get brought into the cell.
Each organelle has ___ proteins to ____
Each organelle has unique proteins to help aid its function.
How do organelles get to these unique proteins?
1. Gated transport
2. Vesicular transport
3. Protein translocation
Rely on signal sequences and recognition by sorting receptors!
when is gated transport used
when is vesicular transport used
when is protein translocation used
Protein Translocation
Process of a protein moving across a membrane.
How do proteins translocate to the correct organelle?
Proteins contain short continuous sequences of amino acids that determine destination of protein and act as signal sequences for their translocation.
Vesicular transport:
Transport of proteins from one compartment to another by membrane-bound intermediates (vesicles)
Vesicle
a membrane enclosed dynamic cellular organelle that is involved in the selective transport of proteins/materials to other compartments
Each signal sequence specifies a particular destination in the cell.
A. True
B. False
a
what is Proteins made in cytosol =
Translated on “free” ribosomes in cytosol
what are some proteins made is cytosol
• Cytosolic proteins = many enzymes, cytoskeletal proteins
• Membrane-associated proteins
• Proteins that reside in and on surface of nuclei, chloroplasts, & mitochondria
• Proteins that reside in peroxisomes
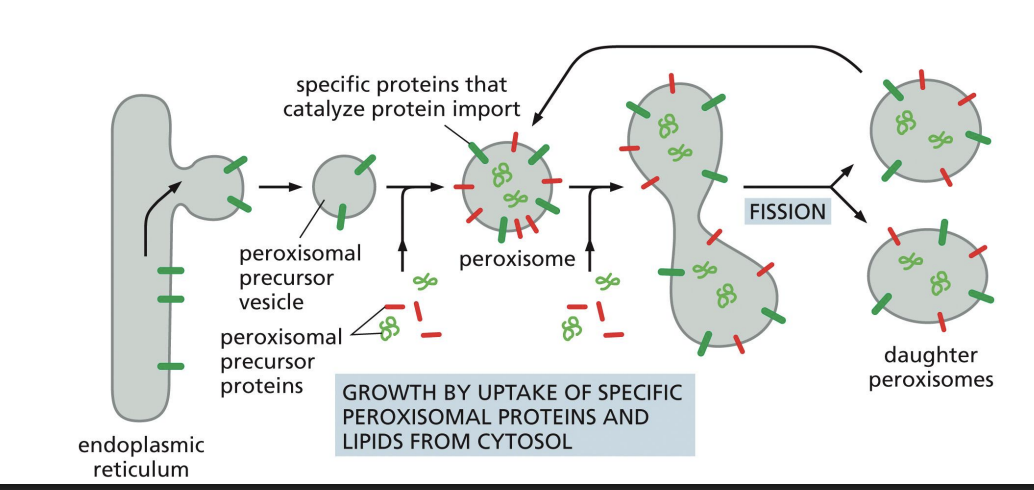
Peroxisomes
• Simple membrane-bound organelles
• Dense core of enzymes involved in many processes
• Site of synthesis and destruction of hydrogen peroxide (H2O2)
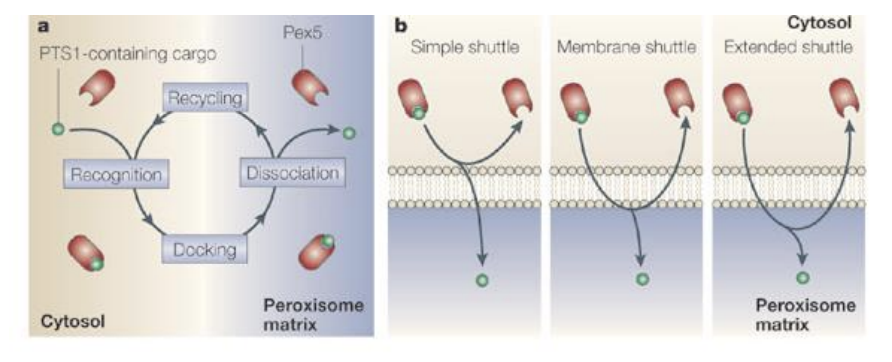
Peroxisomal Protein Translocation
Soluble peroxisomal proteins are made on free ribosomes in cytosol and have signals that receptors (Pex5) recognize to direct transport to peroxisome.
PTS 1 signal = “ser-lys-leu” at C-terminal end of protein.
Membrane Peroxisome Proteins
Peroxisome precursors bud from ER with some peroxisomal membrane proteins that contain a membrane Peroxisomal Transport Signal (mPTS).
can go into fission and produce daughter
membrane protein comes from er and lipids from cytosol
Nucleus
2 membranes
2 aqueous compartments
Perinuclear space is continuous with ER lumen.
Nuclear pores
mediate protein translocation between cytosol and nucleus.
Nuclear Pore Complex (NPC)
20 different proteins that come together as complex of 500- 1000 subunits
nuclear localization signals
Nuclear proteins have nuclear localization signals (NLS).
Can be found anywhere in protein sequence.
if mutated wont move properly
Nuclear Transport Receptors
recognize nuclear localization signals (NLS). Import = nuclear import receptors.

Import receptors
are typically in the cytosol.
Import receptors can bind the NPC which allow it to pass into nucleus.

Ran-GAP and Ran-GEF
Ran-GAP = cytosol and Ran-GEF = nucleus.
These proteins regulate import receptor and cargo interactions.
Ran-GAP and Ran-GEF process
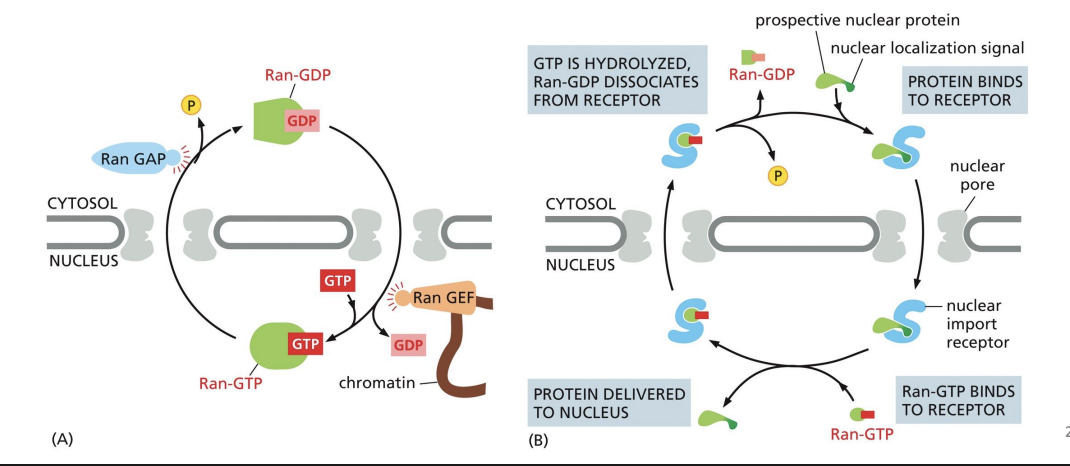
How is it that proteins possessing different kinds of nuclear localization signals (NLSs) can be efficiently transported into the nucleus through nuclear pore complexes (NPCs)?
A. The NPC can bind various NLSs through its unstructured FG-repeat domains.
B. There are specialized NPCs that bind each of the different classes of the NLSs.
C. Different import receptors recognize different NLSs and then interact with NPCs.
D. Proteins with different NLSs travel through different sub-channels within NPCs.
c
The endomembrane system
also called the secretory pathway. (this patthwasy allows to send things outside of plasma membrane )
Proteins move from ER to Golgi to the outside of the cell.
structure of Endoplasmic Reticulum (ER)
The ER is structurally and functionally diverse and is the “early” secretory pathway organelle. (first thing to pick things up )
A system of membranes that encloses a space (“lumen”) that is separated from the cytosol.
2 sub-compartments: smooth ER (SER) & rough ER (RER)
RER
• Studded with ribosomes
• Synthesis of secretory pathway proteins & some lipids
• Site of protein folding & modification
SER
• Synthesis of steroid hormones (gonad and adrenal cells)
• Storage of calcium ions (triggers muscle contractions)
• Detoxification of alcohol & barbiturates (liver cells)
Proteins made in the ER = translated ___
Proteins made in the ER = Translated on ribosomes attached to RER
what are some proteuns made in the ER
• Secreted proteins = hormones, neurotransmitters
• Transmembrane proteins
• Soluble proteins that reside in the endomembrane system organelles e.g. ER, Golgi, lysosomal, vacuolar proteins
Signal sequences that target proteins to the ER generally:
• Are relatively short: ~15-40 amino acids
• Contain non-polar stretch: ~8 or more nonpolar amino acids at its centre
• Are typically at the N-terminus of proteins
Endoplasmic Reticulum (ER) Protein Translocation
Translation begins on free ribosomes and only after the signal sequence is translated can the whole complex move to the ER membrane.
SRP targets the ribosome and partially synthesized polypeptide to the ER.
SRP binds to an SRP receptor on the ER associated with the translocon
Once ribosome docks to translocon, SRP is released and translation resumes.
SRP Mediated Targeting
SRP interacts with SRP receptor on ER membrane.
Docks free ribosomenascent protein complex at the RER translocon.
Chaperone Proteins “Plug” Translocon

Once ribosome is docked at translocon
SRP is released from its receptor
The signal and nascent protein interact with the translocon = translation resumes
Once nascent protein interacts with translocon and translation resumes:
Chaperone proteins (plug) in the ER aid in the entry and folding of nascent protein = translocation
Upon translocation into the ER:
Signal peptidase removes the signal from the N-terminus of the nascent protein and translocation continues
Model for Synthesis of Soluble Proteins in Secretory Pathway

Which of the following correctly describe steps required for protein transport into the rough ER? Select all that apply.
A. Passing of the protein to a protein translocation channel in the ER membrane
B. Initial transfer of the signal sequence to the inside of the rough ER
C. Recognition and binding of the protein signal sequence by SRP
D. Cleavage of the signal sequence from the protein by signal peptidase
a,c,d
ER Protein Co- and Post-Translation Translocation
RER can support both co-translation translocation and post-translational translocation.
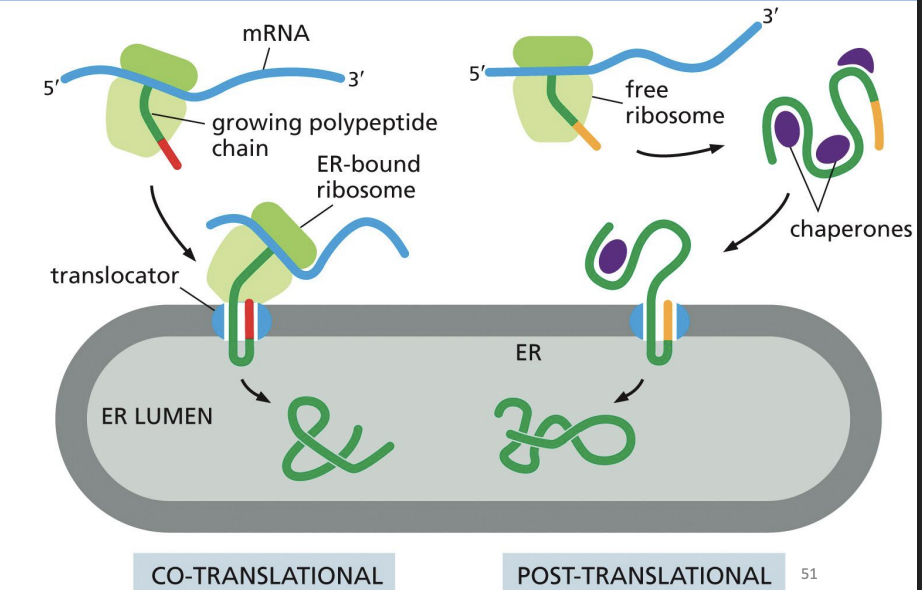
ER translocons are structurally ____
similar
ER Protein Post-Translation Translocation
Some proteins do not enter ER during translation.
Still need signal sequence to translocate
ER Membrane Protein Translocation
Membrane proteins for any secretory pathway organelle must first enter ER. Proteins have hydrophobic “stop-transfer” sequences that stay in membrane
Orientation of transmembrane regions is based on charge (cytosolic accommodates + charge and luminal end accommodates - charge).
Some proteins have ____ transmembrane domains.
multiple
ER Membrane Protein Translocation - Proteins with Cterminal signal sequence
Proteins with Cterminal signal sequence cannot be detected by SRP co-translationally. Post-translational detection by Get proteins.

ER Membrane Protein Translocation - Proteins containing C-terminal signal sequence
Proteins containing C-terminal signal sequence direct addition of a lipid anchor = glycosylphosphatidylinositol (GPI) anchor.

So, if you are a soluble secretory pathway protein, you have completely entered the ER lumen and if you are a membrane protein, you have been inserted into the ER membrane. Now what?
Proteins in the ER have to:
- Undergo folding to form secondary, tertiary & quaternary structures; form disulfide bridges
- Be modified
- Be recognized for transport out of the ER (sorting based on signals)
ER Protein Folding
A multitude of chaperones assist in the folding of proteins in the ER e.g. BiP, GRP94, Calnexin (CNX) & Calreticulin (CRT)
Note that PDI (Protein Disulfide Isomerase) assists in the formation of disulfide bonds and is therefore also required for proper folding of proteins
ER Protein Modification
As proteins fold in the ER, enzymes can modify them as part of their maturation process.
The most common modification of proteins is Glycosylation
Glycosylation
addition of sugars (carbohydrates) onto the proteins.
Nearly all proteins made in the ER are modified co-translationally by glycosylation
whys is Glycosylation imp
• Helps proteins to fold to their final (native) form
• Aids interaction with other macromolecules
Glycosylated proteins
glycoproteins
Oligosaccharyl Transferase
Enzyme that adds the sugars on the proteins
N-linked glycosylation
N stands for asparagine and the sugar chains are added to asparagine in a protein that is located in a motif: Asn-X-Ser/Thr
N-glycosylation occurs as follows:
1. Sugars are built on a lipid called dolichol on the cytosolic face of the ER membrane
2. Dolichol then flips over into the lumenal side
3. The sugars are then transferred onto the protein
What if proteins don’t fold properly in the ER?
If misfolded proteins were to get out to their final destinations in the cell, they could be really harmful.
For example:
• Interfere with other proteins
• Activate wrong signals
• Make protein aggregates that clog up organelles
The ER therefore has a very strict “quality control” system in place.
• Checks to make sure proteins have folded properly
• Allows time for hard to fold proteins to reach their final form
• Destroys proteins that are terminally misfolded
ER Protein Folding: Calnexin Cycle
1. N-linked sugar is added to protein.
2. Two glucoses are trimmed by glucosidases I and II from the glycoprotein
3. Monoglucosylated protein binds to Calnexin (chaperone) which helps the protein achieve its native conformation.
4. Glucosidase II then trims the final glucose from the protein that has had some time to interact with Calnexin • Trimming of the final glucose releases protein from Calnexin
5. If it is properly folded, then it is allowed to exit the ER.
6. If it is improperly folded or unfolded, then GT adds another glucose
7. The re-glucosylated protein re-enters the calnexin cycle - gets another chance at folding
8. If it folds correctly, it can exit.
If the protein remain unfolded or improperly folded for several cycles, then the protein is deemed defective and destroyed (degraded).
9. The protein to be destroyed is moved from the ER to the cytosol where the proteasome degrades the defective protein.
Which of the following correctly describes the sequence of steps in the calnexin cycle during the quality control of N-linked glycoprotein folding in the endoplasmic reticulum?
A) Glucosidase I removes one glucose → Glucosidase II removes the second glucose → Calnexin binds the diglucosylated glycoprotein → UGGT removes mannose residues to retain the protein in the ER.
B) Glucosidase I removes one glucose → Glucosidase II removes the second glucose → Calnexin binds the monoglucosylated glycoprotein → Glucosidase II removes the final glucose once properly folded → If misfolded, UGGT adds back a glucose residue to restart the cycle.
C) Calnexin binds the fully glycosylated glycoprotein immediately after transfer → Glucosidase I removes all three glucose residues → UGGT adds glucose residues → The protein is exported regardless of folding status.
D) Glucosidase I removes all three glucose residues → Calnexin binds the unglycosylated protein → If misfolded, UGGT adds a mannose residue to mark it for degradation.
b
How does the ER switch from the calnexin cycle to extraction and proteasomal degradation?
A slow acting mannosidase removes a mannose from this N-glycan sugar tree = signal for extraction from ER (recognized by the Lectin)
Imagine a protein that has been engineered to contain a nuclear localization signal, a C-terminal peroxisomal targeting sequence, and a standard endoplasmic reticulum (ER) signal sequence. With all of these signals, where would you expect to find the protein after its synthesis?
A) Cytosol
B) Nucleus
C) Endoplasmic Reticulum (ER)
D) Peroxisome
c
According to the model for nuclear protein import, what do you think would happen if you could artificially limit all Ran-GAP activity to the nucleus and all Ran-GEF activity to the cytosol?
A) Proteins containing a nuclear localization signal (NLS) would be actively exported from the nucleus.
B) Protein import to the nucleus would stall.
C) Protein import would occur more rapidly.
D) Nothing would change as this is the usual distribution for Ran-GEF and Ran GAP
a and b
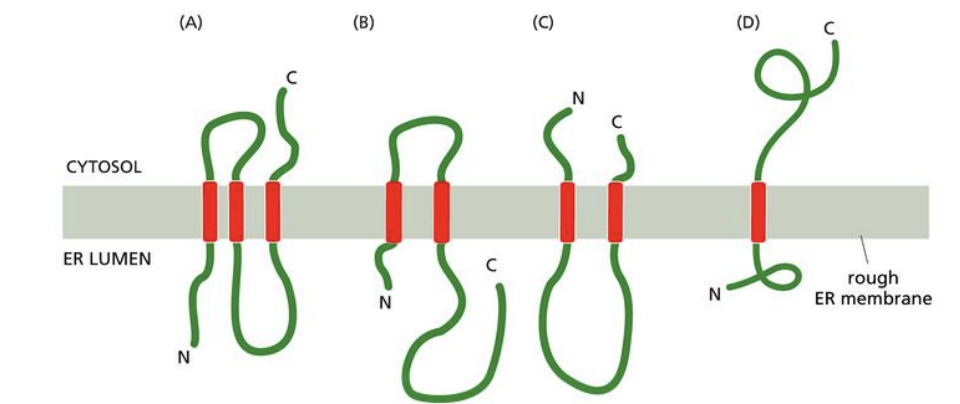
A certain membrane protein is found on the ER membrane with the N-terminus on the ER side of the membrane and the C-terminus on the cytosol side of the membrane. The protein contains three transmembrane domains. What would be the orientation of the protein across the ER membrane if the gene were mutated such that the third transmembrane domain coded for hydrophilic amino acids?
A) A
B) B
C) C
D) D
b