IMED1002 - DNA (I and II)
1/30
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
31 Terms
Abbreviation for nt
nucleotide
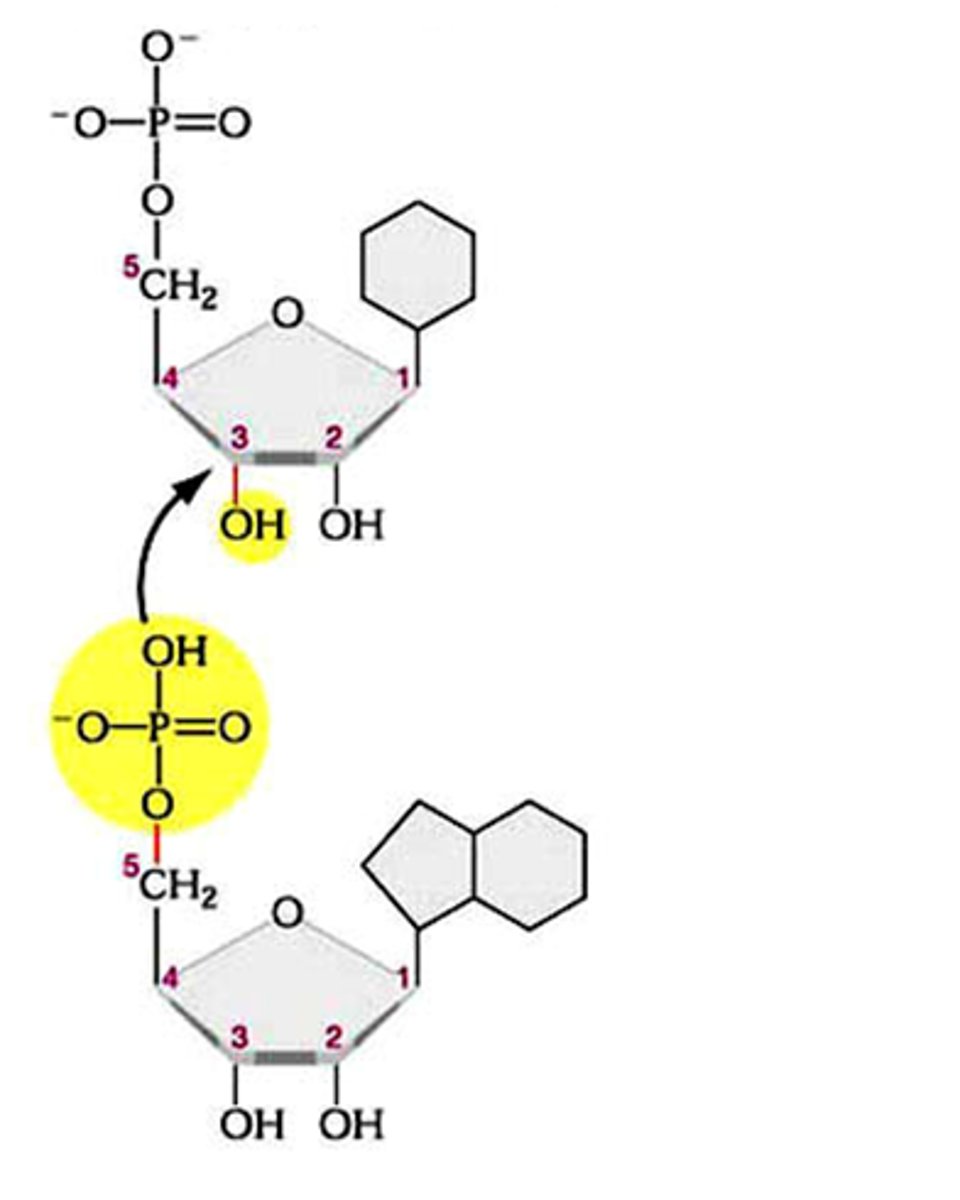
Phosphodiester bond
the type of bond that links the nucleotides in DNA or RNA. joins the phosphate group of one nucleotide to the hydroxyl group on the sugar of another nucleotide
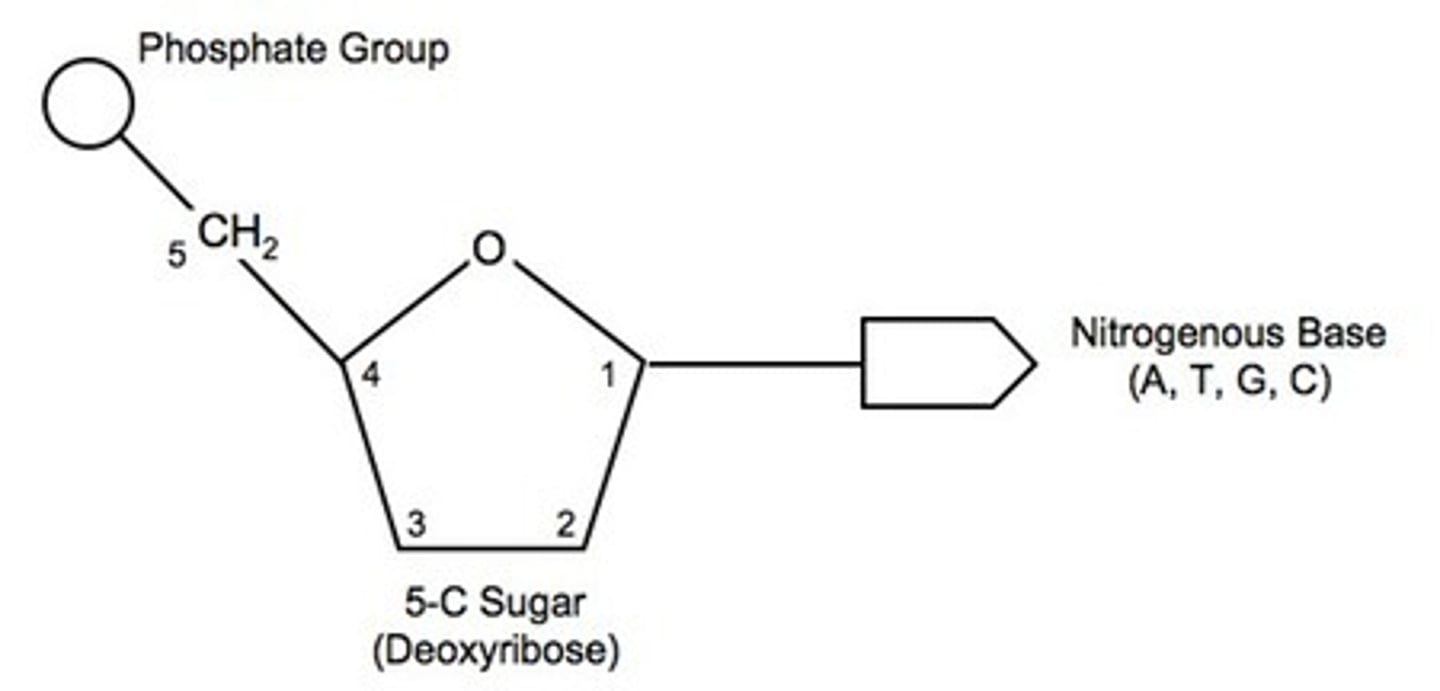
DNA and nucleotide structure
- Sugars' Carbons are designated 1', 2', 3', 4', 5'
- Base joins to the 1' carbon
- Phosphate to the 5' carbon
- Next nt to the 3' carbon
- So DNA has a 5' and 3' terminus (like proteins have ends)
- in DNA, ribose lacks O at 2' carbon, hence deoxy
DNA - a polymer of deoxyribonucleotides
- phosphate groups "bridge" 3' OH of one nt to 5' C of next nt = phosphodiester bond
- phosphate groups ionised so negative charge at physiological pH (DNA is an acid)
- Covalent "Backbone" of DNA is hydrophillic. Consists of alternating sugar-phosphates
Conventions for writing NA (nucleic acid) sequences
- Each base is symbolised by first letter, A, C, G, T (U)
- Phosphate groups are symbolised by ℗ (P in a circle)
- Always write NA in 5' to 3' direction
Actual Structure of DNA (Primary, Secondary etc.)
- like protein structure, it is useful to describe NA structure in terms of hierarchal levels of complexity
- Primary Structure: sequence of nts from 5' to 3' e.g 5' ATCGTTTACAT 3'
- Secondary Structure: involes two strands running antiparallel, development of model explains Chemistry, Physical measurements and DNA's ability to carry genetic information
Chemistry of DNA Structure
1. Hydrophobic bases, & hydrophilic -vely charged backbone of phosphates & sugars
2. A segment of DNA contains the same amount of phosphate & pentose, but the number of individual bases differs (base sequence varies).
3. Exact base composition varies with species.
4. Regardless of the species, in DNA:sum purines = sum pyrimidines, A+G = T+C & amount of A = amount of T, & C = G.
5. Base composition of DNA from different cells in a specimen is identical.
6. Base composition does not change with age, nutritional status, environment.
- (all somatic cells have same DNA in all organisms)
- notice that big (pyrimidine) and small bases (purine) go together. e.g Thymine (Purine) and Adenine (Pyrimidine) go together, as well as Cytosine (Purine) and Guanine (Pyrimidine)
- therefore he realised that there was pairing of bases equidistant
Experiments that showed that DNA carried info from one cell to the next cell
TWO EXPERIMENTS:
GRIFFITH 1928 (cell transformation):
- watch a video on yt coz its confusing to explain
AVERY, MACCLEOD AND MCCARTHY 1944
- watch vid coz its confusing (basically it shows that DNA can transform extracts into a fully functional form, in this case it was a bacteria that killed a mouse)
Model of DNA as a double helix
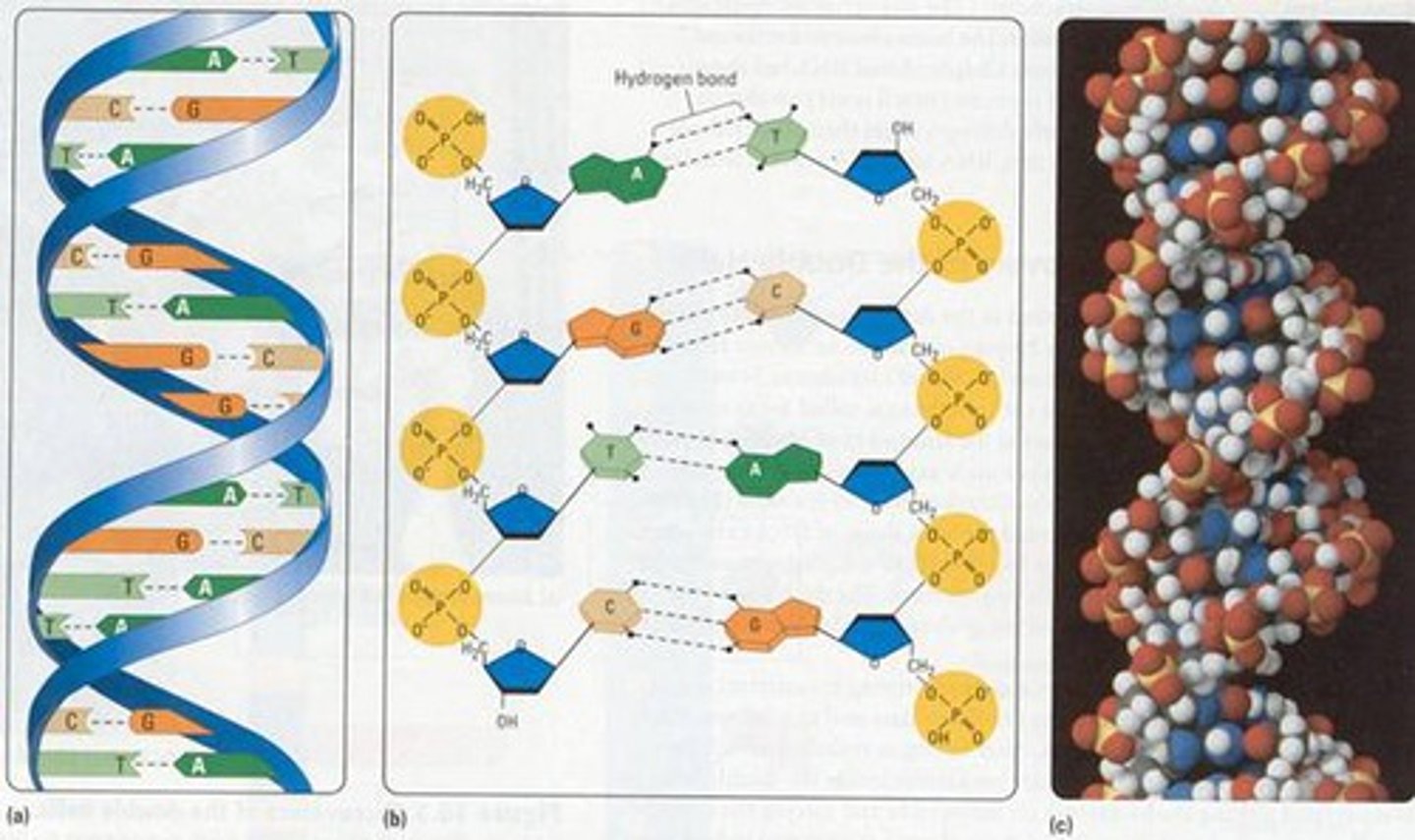
- two nucleotide chains could associate by H bonding between base pairs
- Hydrogen bonds: hydrogen atom shared between two other atoms, both electronegative (such as O and N)
- great diagram on slide 14 and 15 of Lecture 8 - DNA 1
- hydrogen bonds are strongest when three atoms are in a straight line
- bases in DNA hydrogen bond to form specific pairs known as complementary base pairs (A with T, C with G)
- great diagram on slide 16 on Lecture 8 - DNA 1
Why other pairings dont work
- great diagram in slide 17 on Lecture 8 - DNA 1
- shows that while H bonds can be formed between noncomplementary bases (e.g Guanine and Thymine) it is much weaker then complementary bases
Hydrophobic interactions in DNA
- double bonds of bases make pyrimidines and purines (nearly flat) planar
- in water, hydrophobic groups cluster together (hydrophobic interactions)
- these hydrophobic interactions (stacking) as well as dispersion and dipole dipole interactions stabilise 3D structure of nucleic acids
- stacking minimises contact with water
- H bonds between complementary base pairs also stabilise DNA double helix
OVERALL SUMMARY Of DNA Structure
- sugar-phosphate backbone is on the outside (proteins interact)
- bases project into the inside of the double helix
- the two strands are held together by many hydrogen bonds and hydrophobic interactions between bases
- orientation of the two strands are anti-parallel: 5' to 3' and 3' to 5': directions are opposite
- strands are complementary (C with G, A with T)
- base pairing a purine always with a pyrimidine, i.e large with small.
- specificity of base pairing permits duplication of genetic information by synthesis (DNA replication)
Central Dogma
theory that states that, in cells, information only flows from DNA to RNA to proteins
- DNA --> RNA --> proteins
- proteins contain S (Cys), DNA contains P (in the phosphate)
Viral DNA and RNA
- found in the Hershey-Chase experiment (1952), used T2 page was 50% protein, 50% nucleic acid, that phage reproduce by attaching to bacterial cell and the progeny are found within the bacteria
- showed that DNA contained genetic info not proteins
Why Bases are on the inside
- bases are hydrophobic
Forces holding together the helix
- Held together by two main sets of forces:
- H-bonding between base pairs and hydrophobic "stacking" interaction of bases
- Specificity that maintains base sequence is contributed entirely by H-bonding between complementary base pairs.
- base stacking increases DNA stability
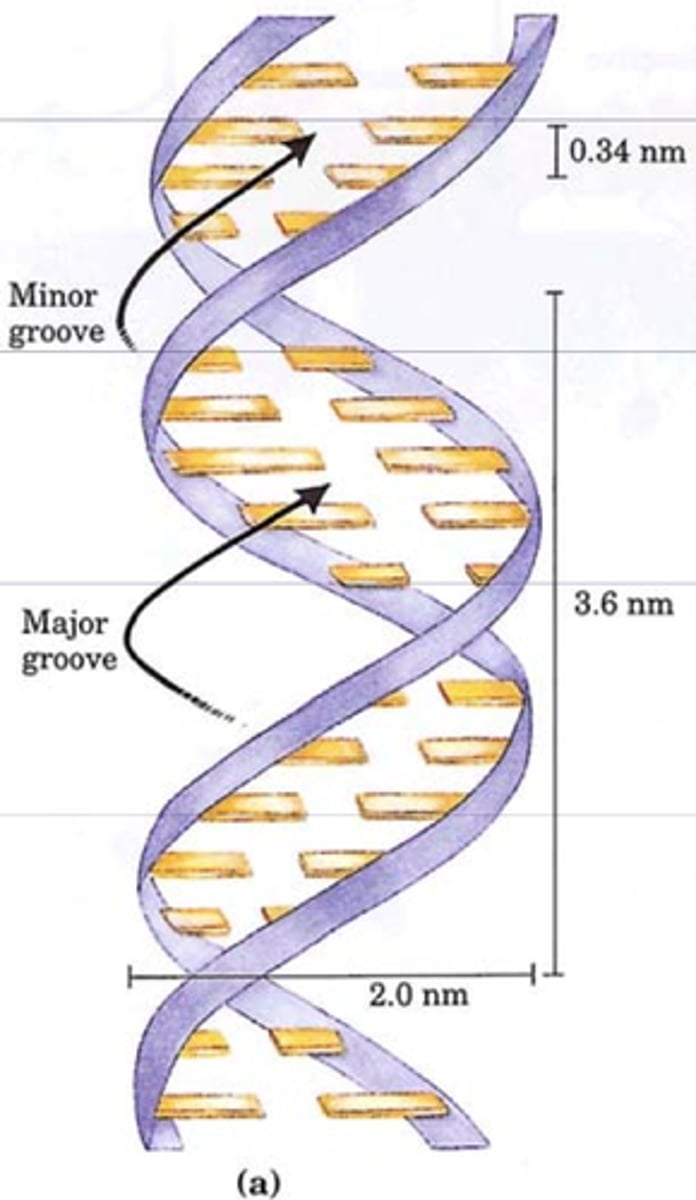
- 10 bases make a complete turn in the Watson-Crick model
- right handed helix with a major and minor groove
Watson-Crick model
the double-helix structure of the DNA molecule
- has minor and major groove
B form
right handed double helix, hydrated (standard form)
- most stable form of DNA under physiological conditions
- two other variants have been identified in the lab, A and Z forms.
- A form is favoured in soolutions devoid (almost) of water, so not our cells
- Z form is a "zigzag", there is evidence short tracts of DNA can take on this shape in cells.
Reversible Denaturation of DNA
- called renaturing/annealing
- H-bonds and hydrphobic interactions are disrupted.
- DNA helix unwinds into 2 strands (no covalent bonds broken)
- Is reversible, especially if over 10 bases are still held together, unwound segments spontaneously rewind = reanneal
Hybridisation
- can have different DNA samples annealing, depending on similarity of primary structure.
- hybridisation is this process done with short DNA sequences or even RNA.
Link between Denaturation and Viscosity
- solutions of carefully isolated, native DNA are very viscous at pH 7 at room temperature
- extremes of pH or elevated temperature results in denaturation
- denaturation decreases viscosity, which can determine the percentage denaturation by studying change in viscosity
DNA Hypo/er-chromism
Single-stranded (SS) DNA absorbs light more effectively than double-helical (DS) DNA
Can determine % denaturation of DNA by looking at change in A260. As DNA denatures, get increased absorption = hyperchromic shift.
Melting Temperature of DNA
- DNA denatures at characteristic temperature
- Tm, melting temperature, is the temperature at which DNA has reached half total maximum denaturation
- can measure this by decrease in viscosity and increase in UV light absorbed (hyperchromic effect)
Number of Hydrogen bonds between bases
- C and G are held by 3 hydrogen bonds
- A and T are held by 2 hydrogen bonds
Differences in Melting Temperature DNA
- Tm variation between two different DNA preparations reflects base composition and length of DNA = differences in forces holding the two sets of DNA strands together
- since G-C has 3 bonds, A-T has two bonds, as G-C content increases, Tm increases
Factors Affecting Melting Temperature (Salt)
- GC content: as GC content increases, Melting temperature increases
- Salt (ionic strength): concentration of ions in the solution will affect stability of DNA (as concentration of salt increases, it has stabilising effect)
- Ions, such as Na+, will interact with negative charges on phosphate backbone of DNA, this reduces the repulsion between the negatively charged phosphates (major force in destabilising the double helix), thus stabilising the duplex structure
- therefore DNA in water denatures at 20C
- DNA in 0.15M salt denatures at T>20 higher
Factors affecting Melting temperature (pH)
- ionisation state of molecules depends on pH
- decrease pH means protonisation.
- high pH leads to deprotonisation
- large changes in protonisation state leads to loss of H-bonds between complementary strands
- very low pH can cause breakage of glyocosidic bonds
- hence mild alkaline solutions are preferred for DNA denaturation
Factors affecting Melting Temperature (Solutes)
- solutes (italics) in vitro (end italics) that can form H-bonds also lower Tm (decrease stabiliity) of double helix
- organics like formamide and urea lower denaturation temperature and prevent reannealing on cooling by forming H-bonds with the bases and thus not allowing complementary base pairing
Hairpin (DNA)
- forms when a single DNA (or RNA) strand is involved in the base pairing
- isolated DNA solutions containing such sequences can form complex structures with multiple hairpins
Palindromes in DNA
read the same in both directions
Differences between DNA and mtDNA
- long double helices of DNA wound up and around proteins (containing lots of basic AA) to allow packaging into the nucleus
- Mitochondrial DNA is also double stranded but mtDNA is circular
- DNA can: copy itself (replicate), be transcribed into RNA. In the S phase, DNA replication occurs