BMS1062 - W9: Regulation of Gene Expression
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
22 Terms
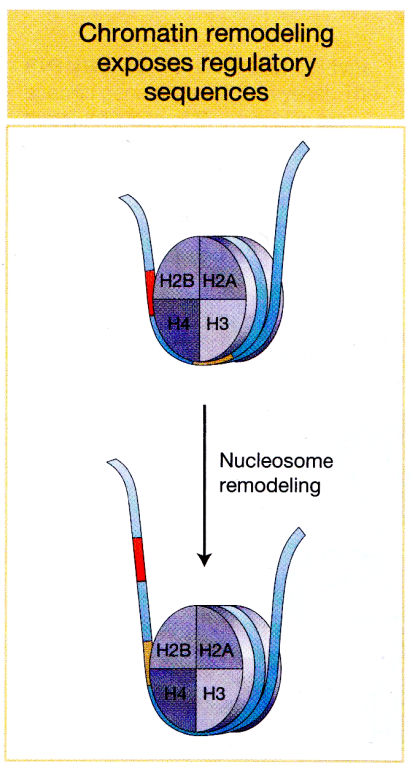
Chromatin Remodeling:
Gene expression can be regulated by having promoters locked in nucleosomes
Chromatin remodeling:
nucleosomes would need to be moved around to free up the promoter region.
changing where the nucleosomes are
how tightly compacted the DNA is around the nucleosomes
can increase or decrease compaction (make genes less expressed or more expressed)
when chromatin is condensed, DNA normally wrapped around histones in a way that promoter region tightly associates with histone proteins. So general transcription factors and RNA polymerase II would not be able to bind to this site
What are some ways eukaryotic cells can change chromatin structure?
roll the entire nucleosome along DNA so promoter region becomes exposed
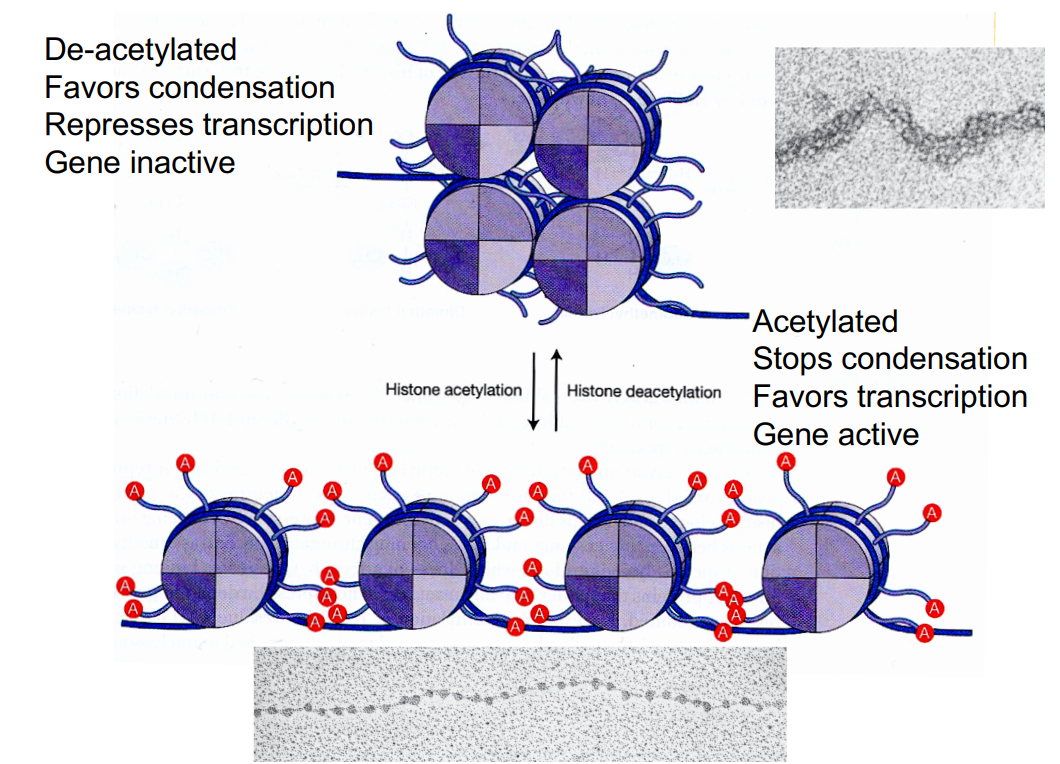
Modification of histones:
Histone tails can be acetylated or methylated → which controls gene expression via different mechanisms.
Acetylation of histone tails:
Histone tails can bind next nucleosome and adjacent DNA
Histone proteins have N-terminal ‘tails’ that stick out of the nucleosome
these tails contain many positively charged lysine residues
nucleosomes have grooves on surface that are negatively charged
so tails of histones of one nucleosome can lodge themselves into negatively charged grooves on surface of next nucleosome, associating with each other.
positive histone tails will also form favorable electrostatic interactions with negatively charged phosphate backbone of the DNA.
Acetylation (CH3CO) of lysine on histone tails stops interactions with neighboring nucleosome and DNA (by removing positive charge). This prevents nucleosomes from condensing, favoring gene transcription.
Histone acetyltransferase (HAT): catalyses acetylation
enzyme that transfers an acetyl group (from acetyl-CoA) onto histones
Loose HAT (Travis’ way of remembering - HAT makes chromatin loose = favor gene expression)
Histone deacetylase (HDAC): catalyses deacetylation
enzyme that removes the acetyl group from the histone tail
Levels of histone acetylation:
more acetylation = more transcription
less acetylation = less transcription
Some eukaryotic activator proteins direct acetylation of histones near promoter.
opens chromatin structure and increases gene expression
function as activators because they promote acetylation of histones near promoters
Some eukaryotic repressor proteins direct deacetylation of histones near promoters
- compress chromatin strcuture and reduces gene transcription
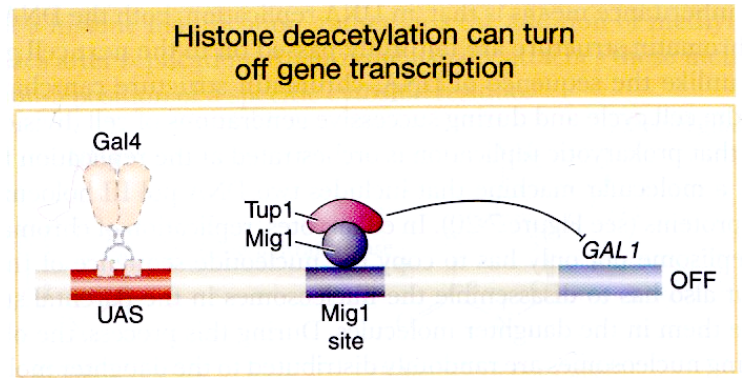
Histone deacetylation in GAL:
In the presence of galactose, the activator Gal4 is active (as galactose is bound)
However, in the presence of galactose and glucose, transcription of GAL1 is repressed regardless of the presence of the activator Gal4 bound to its enhancer.
Mig1 repressor is de-phosphorylated in the presence of glucose, binds between the enhancer and promoter and recruits Tup1 that has an HDAC activity. Histone tails becomes deacetylated in this region, which turns the gene transcription off
bc glucose is the superior energy source, so the cell only wants to express GAL1 when glucose is absent.
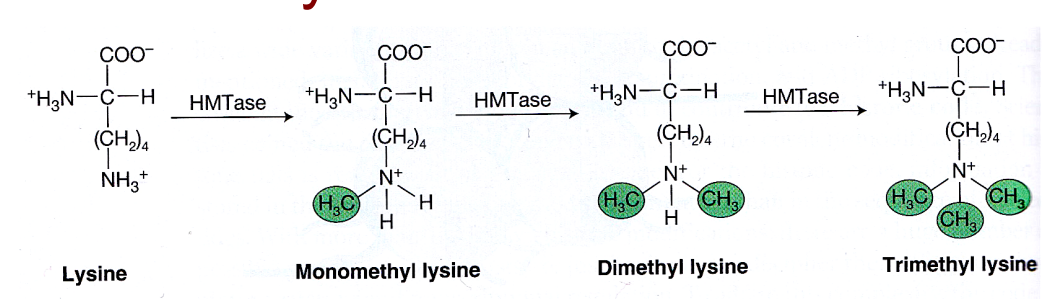
Methylation of histone tails:
both lysines and arginines can be methylated
Histone methyltransferase (HMTase) enzyme catalyses the methylation reaction
1, 2 or 3 methyl groups can be added consecutively to the side chain of lysine. But the positive charge on the side chain of lysine is retained.
so does not affect charge (does not neutralize charge) - so does not directly affect condensation
instead, it creates binding sites for either activator or repressor proteins (depending on the residues modified)
can activate or repress gene expression
DNA methylation and transcriptional control:
DNA can also be methylated
methylation is a covalent modification of DNA rather than histone
DNA is methylated at the 5’ position of cytosines
DNA methyl transferase (enzyme)
GC methylation is associated with inactive genes
This is because DNA methylation leads to transcriptional repression by histone deacetylation
methylation of the cytosine in CG dinucleotides serves as a recognition site for ‘methyl cytosine binding protein’
the methyl cytosine binding protein then recruits histone deacetylase (HDAC) which then removes acetyl groups from histone tails within reach
the CGs are usually around the promoter region?, so it leads to aggregation of nucleosomes = deactivate the gene controlled by this promoter region.
enhanceosome:
enhanceosomes help recruit the transcriptional machinery
Example: β-interferon enhanceosome
multiple transcription factors control expression of β-interferon gene
binding of multiple regulators is cooperative (binding of first protein helps other proteins to bind and stabilizes their binding)
enhanceosome forms binding site for GCN5 (a HAT - histone acetyltransferase)
the HAT causes nucleosome to lose its interaction with DNA backbone, allowing the nucleosome to be rolled - but it needs the help of CBP and SWISNF)
the GCN5 dissociates and leaves a vacant space for CBP (coactivator) to bind. CBP also recruits RNA polymerase II and SWISNF
SWISNF is ATP driven to nudge nucleosome 2 off TATA box, and TAT box becomes accessible to TBP (tatabox binding protein) = starts transcription
epigenetic inheritance
Chromatin structure can be inherited (state of the chromatin - loose, condensed)
during replication, both DNA sequences and chromatin passed down
old histones are re-used by both daughter cells and serve as template to guide modification of new histones
DNA methylation pattern can also be inherited
which promoters are active/ inactive can be inherited
in DNA replication, only parental strand will be methylated
DNA methyltransferase recognises these hemimethylated substrates and methylates the other strand according to the methylation pattern on the parental strand
Lecture 18:
Post-transitional events
Cell identity (what makes cells different when they have the same genome)?
Cell identify if defined by its proteome
they express different RNAs and proteins
Where does most regulation occur (at what level of the central dogma)?
Transcription (73%
post transitional protein folding:
As polypeptide chain emerges from ribosome (N terminus), the protein starts folding onto itself (often before synthesis of whole chain)
energetically favorable for protein to fold onto itself.
folding into compact 3D shape ensures all the amino acid residues that are important for protein activity are positioned where they should be
some proteins need chaperones to help with folding
What are chaperones?
Help unfold and refold misfolded proteins.
there are different kinds of chaperones e.g. HSP60 (heat shock protein 60) there is a cavity inside that hold protein in a favorable environment to promote folding that would not happen spontaneously
What is the difference between misfolded protein and properly folded protein?
How do chaperones recognize misfolded proteins?
Properly folded - hydrophobic amino acids tucked inside
Misfolded - hydrophobic amino acids exposed
chaperones bind to the exposed hydrophobic regions
chaperones prevents aggregation of misfolded proteins (clumping together)
What happens if protein is misfolded beyond repair?
Degradation of misfolded protein by targeted degradation proteosomes
this is another post-transitional mechanism to control proteins
there is competition between chaperone and proteosome - if it takes too long to refold, it will be degraded by proteosome
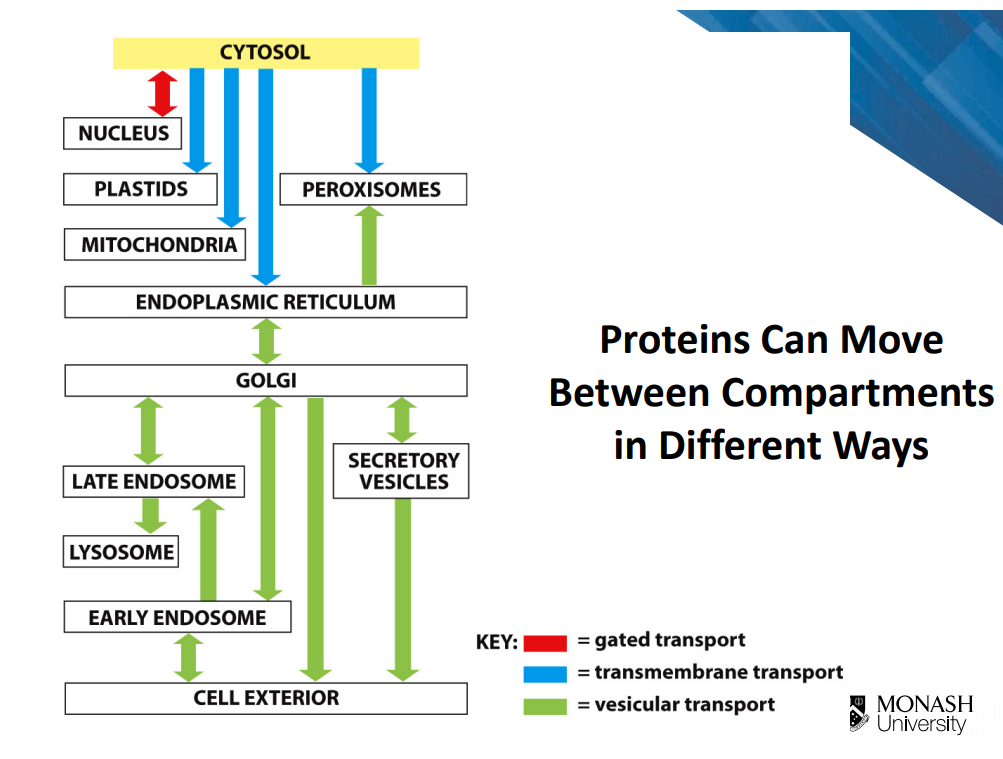
Protein moving between compartments:
Targeting signal/ signal sequences on polypeptide chain
targeted by those signals, proteins can move between compartments in 3 different ways (image)
Gated transport:
Regulated exchange of proteins between nucleus and cytosol
proteins move through pores (but require active transport)
proteins destined for nucleus contain a special nuclear localisation signal (part of polypeptide chain is conserved sequence called nuclear locaisation signal that is recognised by nuclear import receptors
Transmembrane transport:
co-translational translocation
Vesicular transport:
transport via vesicles (budding and fusion)
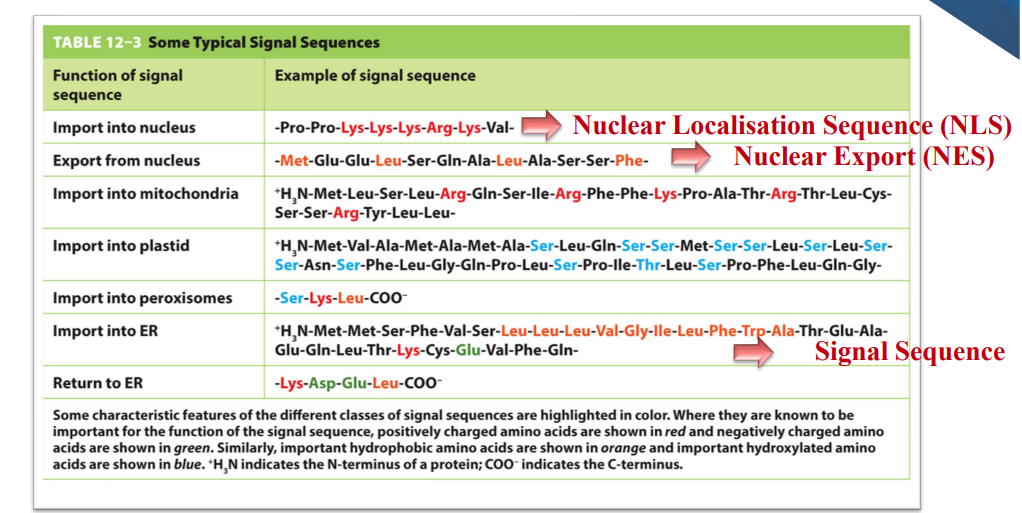
Signal sequences:
sequences are part of the polypeptide chain
orange amino acids = hydrophobic → proteins imported into ER have a stretch of hydrophobic amino acids
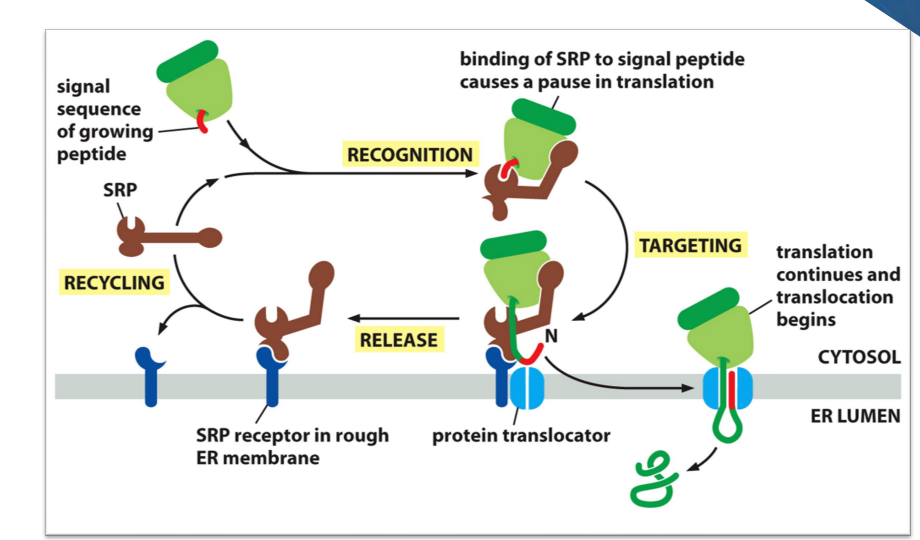
Signal-Recognition Particle (SRP) directs ER signal sequence.
Hydrophobic stretch comes out of ribosome first
SRP binds to hydrophobic stretch and causes a pause in translation and delivers the ribosome to the protein translocator in the membrane of the ER.
beginning of protein then threaded through the channel. As translation resumes, it passes through the channel to the inside of the ER.
After translation finished, it is released to the inside of the ER
SRP dissociates and is recycled
in image, the protein doesn’t have red part anymore, because the signal recognition peptide is cleaved off soon after it enters the protein translocator channel.
Posttranslational modification in ER:
Glycosylated
addition of a common oligosaccharide
covalently attached to side chain of an asparagine residue (N-linked)
may be required for folding, stability and function
glycosylation tegs molecules/ part of molecules that end up going outside the cell
Stablised by disulfide bonds
covalent bond between 2 cysteine side chains
intra- or inter-molecular crosslinks
requires oxidative condition (need oxygen)
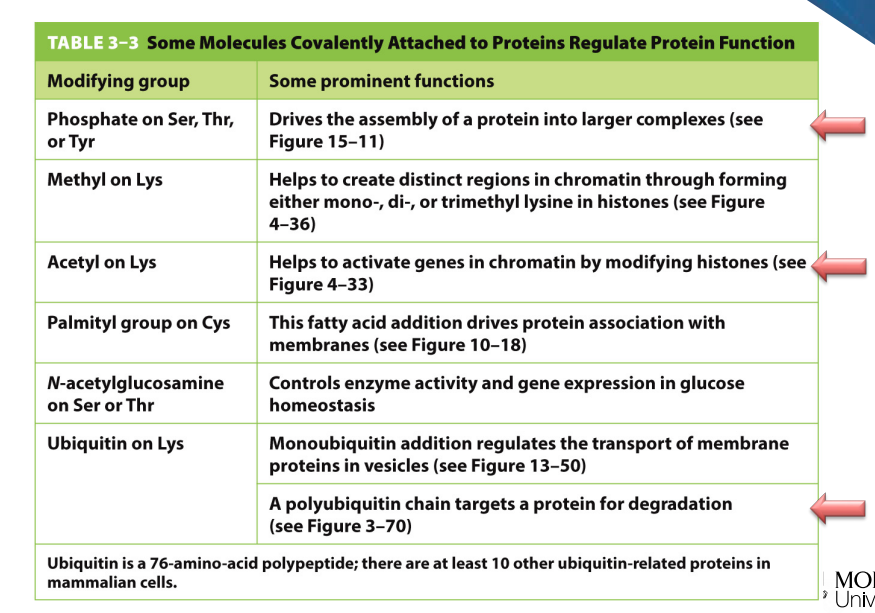
Covalent modifications:
Proteolysis (cleavage pf the protein)
Glycosylation (addition of sugars)
Phosphorylation (addition of phosphate group)
Examples of chimerical modifications:
Phosphorylation on what amino acids?
Methylation on???
etc.
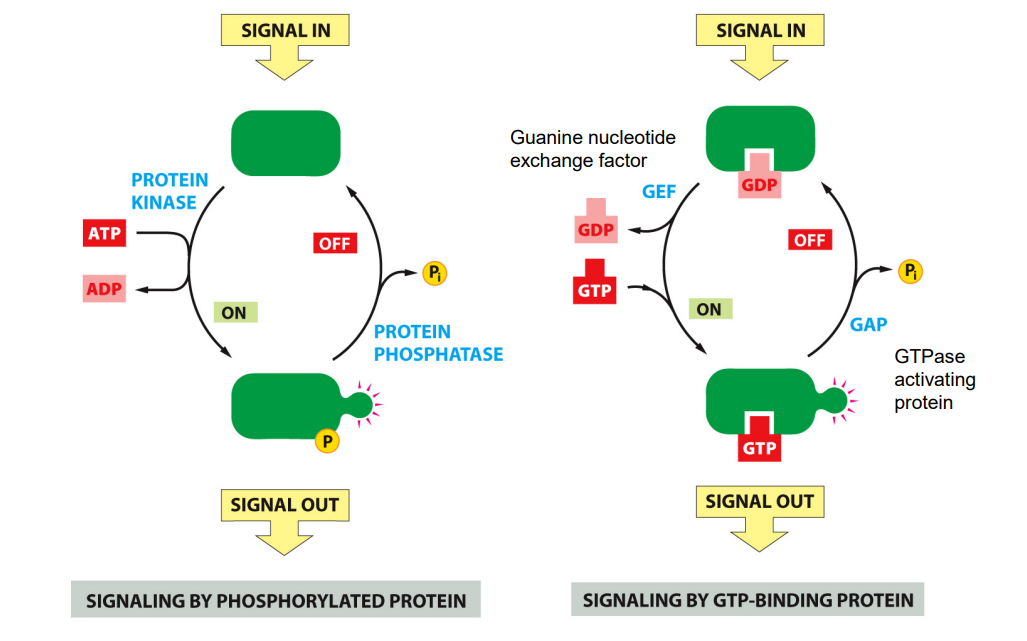
Two ways of activating proteins in cell signaling cascades:
Signaling by phosphorylated protein
phosphorylation is catalysed by a family of proteins known as protein kinase
in response to signal, kinase can transfer a protein group form ATP to a substrate protein which turns that protein on so it can perform actions on other components of chain actions
By phosphorylation is reversible - catalysed by an enzyme called protein phosphatase can remove the phosphate group from these protein, switching it off.
E.g. of signal: growth factor (signals cell to divide)
Signaling by GTP-Binding protein
protein activity is determined by whether a GDP or GTP is bound to the protein
Protein e.g. Rus are activated by GTP and inactivated by GTP hydrolysis
in response to a signal, a protein called Guanine nucleotide exchange factor can promote dissociation of GDP and binding of GTP
Also reversible - protein called GTPase activating protein can promote GTP to GDP, deactivating the GTP binding protein and inactivated