BIOL 2010 - Control of transcription in eukaryotes
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
14 Terms
types of polymerase
POL I
transcribes ribosomal RNA
POL II
transcribes genes that encode proteins and most snRNA
POL III
transcribes tRNA, some RNA and other small RNA
transcription factors:
IMPORTANCE
determine whether transcription occurs
determine cell specificity
confer responses to specific stimuli
ISOLATION OF TFS
to find TFs: bioinformatic analysis of the whole genome and look for conserved regions
STRUCTURE OF TFS
TFs have a modular structure
one region binds to DNA
another region binds to the components
all the AAs for DNA binding is found in one region
oct 2 has 60AAs region which binds to DNA —> this 60AA region has the same affinity as the full length of Oct 2
the other region binds to other things either bound to activation domains or inhibitor domains (sometimes this binding is direct, sometimes through intermediate cofactors)
ISOLATION OF TFS
DNA sequence encoding TF is synthesised and bound to beads
proteins are extracted from lysed cells and combined with the beads
the TF protein will bind to binding sequence so will stick to the beads
protein can then be extracted from the bead
classes of DNA binding domains
most Tfs have 1 of these 3 binding domains
these are different strategies of proteins for inserting themselves into DNA so that they fit tightly
inserted into the major groove due to charge, size, shape etc
zinc finger
helix turn helix
basic binding domains
Zinc finger
protein that contains an alpha helic and Zn2+ which keeps all of the AAs in shape
usually attached in a multimeric way
EXAMPLE: Sp1TF
contains a loop of 23 AAs
link between multiple zinc fingers is 7-8 AAs
alpha helix contains major groove
specifically driven by multiple zinc fingers
Zn2+ doesnt directly interact with the DNA but is essential for the folding of the finger
zinc fingers bind both the major and the minor grooves
Helix turn helix
alpha helix, turn followed by another helix
one is a stabilising helix and the other is a recognition helix
multiple helixes and turns —> most commonly 3
E.g: homeobox TFs
basic binding domain
TFs w/ basic binding domain cant bind alone so they must dimerise
form 2 prongs that can straddle the DNA helix
E.g leucine zipper or helix loop helix
regulation of TFs
may be regulated by location if responsiveness is important
E.g: steroid hormone receptors (they have a zinc finger binding domain)
E.g: vit D receptors
VIT D RECEPTOR
Vit D receptor in the cytoplasm
binding of vit D to receptor causes dimerization and translocation into the nucleus
in the nucleus it binds to a consensus sequence which causes transcription of VD responsive genes
how do transcription factors activate transcription?
binding region alone doesnt cause transcription
activation/repressor domain is responsible for transcription
to determine which region is the activator/repressor you can take a well known TFs binding domain and add on parts of an unknown TF which you suspect may be an activation domain
whichever one causes transcription will be the activation domain
regulation of elongation
regulated by promoters upstream of the core promoter
cell fate determined by gene expression —> regulated by TFs
this control is controlled by tissue specific TFs:
UPSTREAM SEQUENCE ELEMENTS:
can enhance the binding of PIC to increase transcription of a gene
regulator proteins binding affects affinity of RNA pol to promoter
E.g: GC box and CAT box
upstream sequence elements present in housekeeping genes
must be in the same orientation as the gene to be effective
E.g: myo D is a gene found in all cells but is only expressed in muscle cells
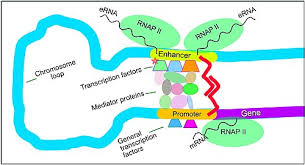
enhancer/activator regions
usually a long way upstream/downstream of the gene
not part of the promoter but works in a similar way
bound by activator proteins
DNA structure means that the enhancers are close to the promoter due to the way it folds
can be any orientation (5’—>3’ or 3’—>5’)
can also be found in the introns so mutations in the introns will still affect the phenotype
different types of activation/enhancer domains
only have generalised properties, a lot less well defined than the categories of binding domains
activation domains interact with a whole host of proteins depending on the function of the Tfs rather than the binding domains which fit a specific sequence of DNA
Acidic AAs
Glutamine rich
proline rich
how do activation/enhancer domains work?
the pre initiation complex is assembled
the general TFs occupy so much space that the activator protein and regulatory proteins are able to bind to structures of DNA on the opposite sides of the promoter
may cause a conf change in the PIC which is more tightly bound which enables transcription
or through the recruitment of coactivators:
coactivators work by interacting with the PIC
sometimes they work by the interaction w the DNA by loosening or opening the chromatin structure to allow the PIC to bind better to the DNA
how do activators alter chromatin structure
co activators can modify histones
histones have 2 domains
globular domain
amino tail domain rich in lysine
if the coactivator has histone acetyltransferase activity then they can neutralise the +ve charge of the lysine which causes the histone to open up as the DNA no longer attracted to the +ve charge of the histone
transcription can occur more readily
E.g glucocorticoid receptor can recruit co activator p300/CBP which has HAT activity
inhibitory domains
not done in whole blocks of genes, only works on an individual gene basis
inhibitory domains bind to DNA in the same way as the activating domains and they block TFs with activator domains from binding
alternatively, they might bind directly to the PIC and reduce the affinity of the PIC for the core promoter
both these methods work by physically getting in the way but they can also work through co repressors:
CO REPRESSORS:
indirect interaction between PIC and the repressor
opposite of the activation —> close the DNA structure by HDAC (histone deacetylase) removing the acetyl group of histone units which restores the positive charge of the histone, preventing transcription
EXAMPLES:
SMRT
forms a large complex w/ RAR or transcriptional repressors
has HDAC capability
tamoxifen
oestrogen is important in breast cancer development
tamoxifen is a drug that enhances the corepressors to the oestrogen receptor —> prevents proliferation of oestrogen dependent cancers
however cancers can become tamoxifen resistant