BIOL 300 - Chapter 17
1/60
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
61 Terms
what is produced in transcription?
many (360,000 mRNA molecules) RNA copies being produced from one DNA template, all of these copies end up being transient and not passed on
how do DNA strands relate to transcription?
one strand is used as a template for a gene (can be top or bottom) - one strand will be the gene, and the mRNA will look EXACTLY LIKE THIS, so the mRNA has to be made from the opposite strand
what is the sense strand?
NONTEMPLATE strand with the same sequence as mRNA
what is the antisense strand?
TEMPLATE strand that is complementary to mRNA, (its what makes mRNA), and is what is read by RNA polymerase
if a strand has these sequences:
template:
5’ ATGCGTGTCGAAACTGTACTG 3’
3’ TACGCACACCTTGACATGAC 5’
sense strand:
what will the resulting mRNA be?
3' UACGCUCUGCUUGACAUGAC 5'
what four sections make up the anatomy of a gene?
promoter/enhancer region, 5’ UTR, gene sequence, 3’ UTR
what is polycistronic mRNA?
single mRNA molecule has multiple ORFS, with each translating into a protein (seen in prokaryotes, not eukaryotes)
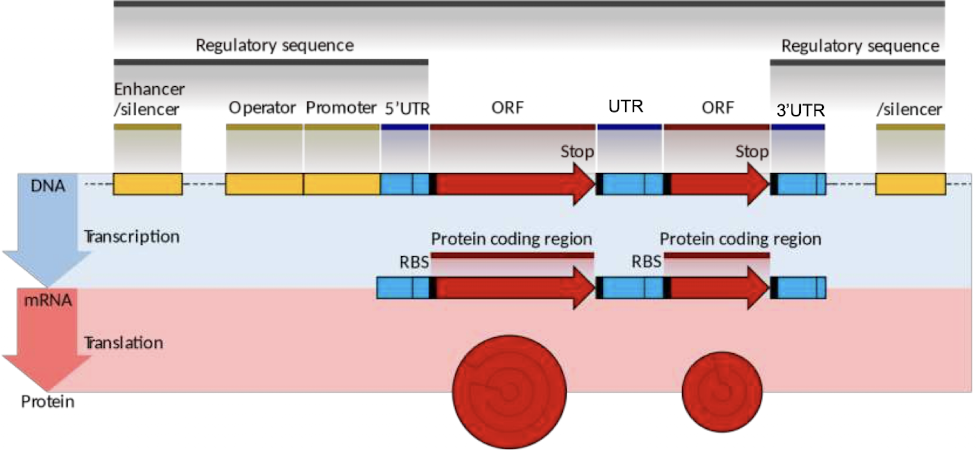
what is an operon?
operating unit (series of polycistronic genes made into polycistronic mRNA) for prokaryotes
what is monocistronic mRNA?
mRNA only codes for one protein product, and only has one ORF - seen in eukaryotes
what are the important elements of the sense strand?
made up promoter, transcription start point, and transcription terminator
what is the promoter in the prokaryotic sense strand?
binding site for RNA polymerase; usually upstream start site (between -1 and -35), and not in mRNA sequence
what is the transcription start point in the prokaryotic sense strand?
1st base transcribed into RNA at +1
what is the transcription terminator in the prokaryotic sense strand?
sequence that triggers RNA polymerase falling off; unique to bacteria
what is involved in prokaryotic RNA chain formation?
synthesis starts within small transcription bubble; DNA strands separate, RNAP reads template 3’ to 5’, and grows RNA 5’ to 3’ at the free 3’ hydroxyl end
how big does the transcription bubble grow to be?
the transcription bubble doesn’t grow, it stays 12-14bp long - DNA duplex reforms and displaces RNA into single strand
how does prokaryotic elongation occur?
multiple RNA polymerases transcribe simultaneously (in a row), and exhibit christmas tree like structures as the synthesized RNA grows - removes NAPs when needed
what is FISH?
fluorescent in situ hybridization - how geneticists can view process of transcription
how does FISH work?
molecules are fluorescent and complementary to mRNA to track how much is expressed - green is the transcript from one gene, and red is a different, with blue being a single nuclei.
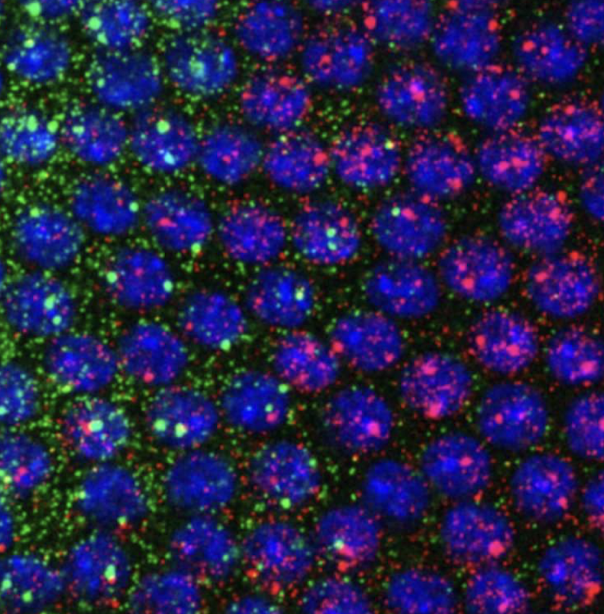
what is the process of visualizing active transcription sites?
MS2 seq repeats are added to genes, and when they’re transcribed, the sequence folds into a loop thats recognized by a RNA binding protein fused to GFP (green fluorescent protein), which makes it fluorescent. fluorescence is proportional to transcript activity, so genes that are expressed more are brighter
how many RNA polymerases do prokaryotes have for mRNA, tRNA, and rRNA?
one
what is mRNA?
messenger RNA - carries DNA genetic code from nucleus to ribosome in cytoplasm
what is tRNA?
transfer RNA - transfers amino acids to ribosome during translation
what is rRNA?
ribosomal RNA - forms structural backbone of ribosomes and has catalytic funciton to help form peptide bonds to link amino acids together
what is the difference between RNA polymerase and DNA polymerase?
prokaryotes don’t have dimer, clamp, or separate exonuclease subunit (has intrinsic nuclease activity, and doesn’t need strict, elaborate proofreading)
what is the structure of RNA polymerase?
two functional parts (core enzyme and sigma factor), and overall made up of 6 proteins, including alpha subunits, beta, beta prime, and omega
what does the core factor do in RNA polymerase?
made up of alpha subunits, beta, beta prime, omega; beta and betaprime are the active site, and alpha and omega are for DNA binding, complex assembly, and interactions w other factors. helps for DNA binding (to any DNA, its not specific)and RNA synthesis.
what does the sigma factor do in RNA polymerase?
helps with promoter recognition, and reduces the non-specific binding of core enzyme to DNA. it makes association constant higher and increases lifetime of binding
is one sigma factor exclusive to one promoter?
no, sigma factors can be used for different promoters
does every promoter have their own sigma factor?
no, one sigma factor can bind to many promoters
how do sigma factors work?
sigma factors can recognize groups of promoters that have similar functions through their consensus sequence
what is a consensus sequence?
calculated order that points out necessity of nucleotides at eah position in sequence alignment - shows most frequent nucleotide at every position in segment
what is a conserved sequence?
a sequence that shares similarities with sequences across species or in different parts of the genome - from duplication and divergence
what is a representative sequence?
group of conserved sequences that involves the nucleotide most likely to be represented
what is an example of consensus sequence binding?
DnaA binding to 9-mers
what are the two most important promoter elements?
-10 element (pribnow/TATA box/first melted seq) and -35 element
what are upstream elements in the promoter region bound by?
alpha subunits, to regulate expression levels
what is sigma factor specificity determined by?
short consensus sequence of each element, and distance between elements
what does a mutation in promoters and sigma factors mean?
alteration of expression of target genes
what could a LOF mutation mean in promoter/sigma factor?
loss of promoter function or deletion of a gene
what does a mutation in a sigma factor mean in terms of gene expression?
expression of all genes controlled by the sigma factor are affected
what does a mutation in a promoter mean?
expression of the gene controlled by the promoter is affected - affects quantity but NOT quality of RNA produced (seq of elements or distance between elements)
can a missense or frameshift mutation occur in promoter?
no, since these mutations occur in coding regions of genes
what would amorphic/hypomorphic/LOF mutations result in inside promoter region?
loss/reduction in expression of controlled gene, or decreased RNA polymerase affinity for the sequence
what would hypermorphic/GOF mutations result in inside promoter region?
overexpression of gene - RNA polymerase won’t leave the gene alone (thats harrassment)
what is sigma factor switching?
genes controlled by a specific sigma factor usually work together
what genes does sigma-70 promote?
genes for exponential growth when there’s enough nutrients
what genes does sigma-32 promote?
genes for a response to heat shock
what determines which sigma factor is predominantly used by the holoenzyme in transcription?
the expression level of the sigma factors - in normal conditions, sigma-70 is dominant and sigma-32 is degraded, but in high temperatures, sigma-32 levels increase and the holoenzyme switches to sigma-32 and predominantly binds to it
can -10 and -35 sequences identify which sigma factor is bound?
yes, they can tell which factor is bound, or which factor will bind
what are the three stages of transcription?
initiation (rate limiting step), elongation, termination
why is initiation the rate-limiting step?
the assembly of different complexes takes up a lot of energy, so if the cell determines it’s not beneficial it won’t complete initiation to save energy
what are the three steps in prokaryotic initiation?
1)form closed complex (duplex DNA interacts w alpha subunit and sigma factor from -55 to +1),
2) complex transitions into open state (DNA and RNAP changes conformation, DNA melts in Pribnow sequence so DNA can go into enzyme active state),
3) complex changes into ternary shape (short RNA chain is made from +1, RNAP is stuck at promoter, but abortive initiation is possible)
which part of initiation is the rate limiting step?
formation of ternary complex, during abortive initiation
what is abortive initiation?
short RNA chain is relased and new RNA chain is synthesized from +1 until RNAP escapes or is released
how does RNAP know how and when to detach during termination?
sequence specific DNA cis element (terminator) tells RNAP to fall off (RNAP doesn’t recognize it)
what are the two kinds of terminators?
intrinsic terminators (doesn’t need other proteins, strong) and rho-dependent terminators
what makes up an intrinsic terminator RNA?
hairpin (stem loop), and U track (downstream from hairpin, forms A-U pair with template DNA)
what is the mechanism for intrinsic terminators?
U track transcription slows down RNAP enough for hairpin to form, and the interaction between the hairpin and RNAP leads to misalignment which leads to weak interaction between u-a termination bond
what is a rho dependent terminator?
weak terminator element that needs help from Rho proteins - alone, it can stall RNAP but not terminate sequencing
what makes up a rho-dependent terminator?
rut site (entry site for Rho proteins and attracts them as a recognition sequence), rho protein (is an ATP dependent helicase with a RNA binding domain)
what is the mechanism for rho-dependent termination?
rho binds to rut site on nascent RNA (newly synthesized RNA molecule that is still in the process of being made), rho tracks onto RNA, rho catches up with RNAP and destabilizes it, rho uses helicase activity to unwind DNA/RNA hybrid to release RNA