Genomics, Proteomics, and Viral Replication Processes
1/116
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
117 Terms
Gene knockdown
- A temporary method to reduce gene expression.
- Uses small interfering RNA (siRNA) or short hairpin RNA (shRNA) to bind to the gene’s mRNA and block its translation into protein.
- It doesn’t change the genome, just prevents the RNA from being used.
- Useful for studying gene function without making permanent changes.
Genome
Complete set of an organism's genetic material.
Genomics
Study of genomes and their functions.
Transcriptome microarray
Measures transcription levels of multiple genes.
how do transcriptome microarrays work
Isolating RNA and reverse transcribing to RNa so it can bind on to the chip
Proteomics
Study of an organism's proteins and their functions
- where proteins are located in the cell membrane
2-D polyacrylamide gel electrophoresis
Technique for separating proteins by size and charge.
- used to study proteins
- each dot represents show amino acid sequence
50% greater sequence similarity
sequence similarity typically have similar functions
70% greater sequence similarity
definitely have similar functions
Metagenome
Total gene content from organisms in an environment.
- microbiomes
Restriction enzyme
Cuts DNA at specific sequences.
- do not pick the resitriction enzymes
sticky ends
Single stranded ends of DNA left after cutting with enzymes
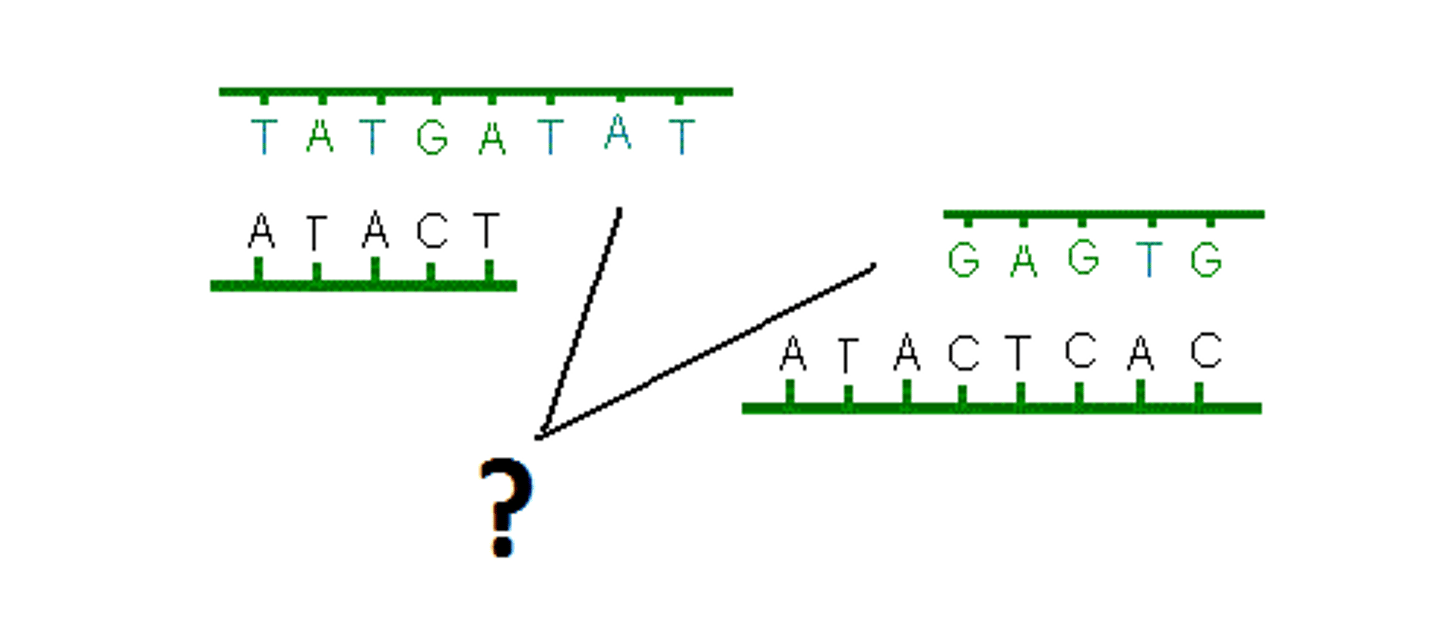
blunt ends
Restriction fragments with no overlapping ends and that never combine with another type of DNA
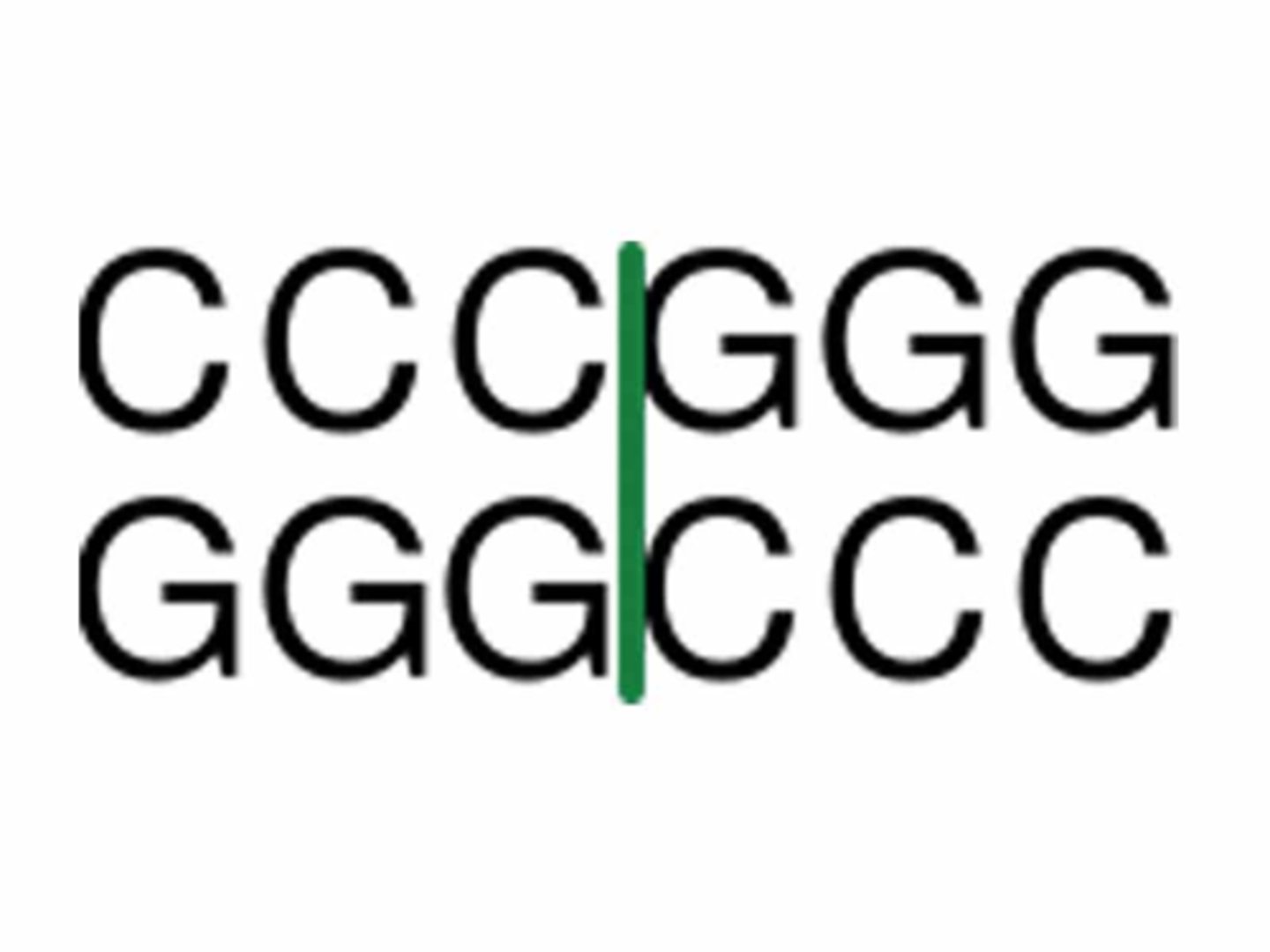
Recombinant DNA
DNA formed by combining DNA from different sources.
basics of a Plasmid vector
Circular DNA that replicates independently in bacteria.
- Replicates indep
- Needs an origin of replication
- Contains a promoter so you can insert your gene
- MCS
- antibiotic resistance
Origin of replication
Site where DNA replication begins.
Promoter
Region that initiates transcription of a gene.
MCS (Multiple Cloning Site)
Region for inserting DNA fragments into a plasmid.
- region where your PCR product gets inserted or gene is going to get inserted
Antibiotic resistance gene
bacteria can survive exposure of the antibiotic
why is the can the antibiotic resistance gene be important in bacteria
we want to select bacteria that constin the vector so use the antibiotic resistance gene in order select for bacteria with the vector of choice gene
Agarose gel electrophoresis
Technique for separating DNA fragments by size.
- Contains ethidium bromide
- Loading dye
- Ladder of known sizes
- "Run to Red"
- Visualize under UV light
Gene cloning basics
1. Foreign DNA
2. Amplify PCR
3. Cut the DNA
4. Make sticky ends
5. Paste with a vector and DNA ligase
6. Introduce to bacteria cells
checking success of a clones
- our vector has antibiotic resistance
- plate on antibiotic its resistant to the only the product desired with grow
telling the plasmid and vectors apart
- blue white screening
- pcr
- restriction enzyme
- sequencing
Blue-white screening
- Method to identify recombinant bacteria colonies.
- looking for white colonies the lac z gene is gone
- blue colonies indicates that the lac z gene is still present
ways to screen for colonies
reliable techniques used to confirm whether your colonies contain the correct DNA insert after a cloning or transformation experiment
1. PCR with primers
2. Restriction ezmyme digestion
3. sequencing
PCR with primers
- use primers specific to your gene of interest.
- If the colony contains the correct insert, PCR will amplify that region, and you'll see a DNA band of the expected size when you run it on a gel
- No band or the wrong size could mean the insert isn’t there or isn't correct.
Restriction enzyme digestion
- isolate plasmid DNA from the colony and treat it with specific restriction enzymes that cut at known sites.
- By analyzing the pattern of DNA fragments on a gel, you can determine whether the insert is present and correctly oriented.
Sequencing
- You isolate the plasmid DNA and send it for DNA sequencing using primers that flank the insert
- This confirms the exact sequence of the insert, ensuring it's correct with no mutations or errors.
how do you confirm that the DNA from the plasmid is being expressed
1) reverse transcriptase PCR
2) western blotting
Reverse transcription PCR
- This checks whether your gene is being transcribed into mRNA, which is the first step of gene expression.
- First, you extract RNA from your cells.
- Then you use reverse transcriptase to convert the RNA into complementary DNA (cDNA).
- After that, you run a PCR with primers specific to your gene.
- If you get a PCR product (a band of the expected size on a gel), it means your gene is being transcribed — a good sign of gene expression at the RNA level.
Western blotting
- This tells you whether your gene is being translated into protein.
- First, you extract proteins from your cells.
- You run them on a gel to separate by size.
- Then you transfer them to a membrane and add antibodies specific to your protein of interest.
- If your gene is being expressed, the antibody will bind and you’ll see a band where your protein should be — confirming expression at the protein level.
size limitations of a plasmid
10,000 basepairs
Homologous recombination
- Adding a gene into a large piece of DNA
- Regions have similar sequences criss cross and swap out to insert the piece of DNA that is synthesized
- Swap out region in the back then recombine
ways to determine whether genes have recombined into the genome
foreign DNA (like a gene you introduced) becomes permanently integrated into the DNA of the host cell—in other words, the new gene is not just floating around in the cell, but actually becomes part of the cell’s genome.
1) replica plating
2) gene knockdown
3) gene knockout
4) CRISPR
5) transposon
6) random mutagenesis
Replica plating
- You grow colonies on a normal plate and then replicate them onto another plate that contains an antibiotic.
- Colonies that grow on both plates are likely unchanged or contain the resistance gene.
- If a colony does not grow on the antibiotic plate, it suggests that a gene important for resistance might have been disrupted by recombination.
- This technique is often used in genetic screens to detect mutations or recombination events.
Gene knockout
- A gene is permanently disrupted (or "knocked out") by inserting a disruptor sequence (like antibiotic resistance or a stop codon).
- This prevents the gene from being expressed or functioning.
- It's often used to study what happens when a gene is missing, helping you understand its role.
- This change is permanent and passed on to daughter cells.
CRISPR
- Uses a guide RNA to direct the Cas9 protein to a specific DNA sequence in the genome.
- Cas9 then cuts the DNA, which can lead to:
- Gene disruption (knockout)
- Insertion of new DNA (gene editing)
- CRISPR allows precise targeting, making it ideal for controlled gene recombination experiments.
Transposon
- A transposon is a mobile DNA element that can randomly insert itself into the genome.
- If it lands in a gene, it can disrupt it, potentially leading to a change in phenotype.
- You can then screen for phenotypic changes to find out which genes were affected.
- Good for random genetic screens.
random mutagenesis
- Introduces mutations randomly in the genome using chemicals, radiation, or transposons.
- You then observe phenotypes and track down the mutated gene responsible.
- Useful for discovering new gene functions or pathways.
Biotechnology
Use of biological systems for technological applications.
1) genetically modified organisms
2) transgenic organisms
3) ecoli recombinant
4) vaccines
5) autrophic ecoli
genetically modified organisms
Genetically Modified Organisms are organisms whose genomes have been altered by the insertion of genes from another organism.
This can involve:
- Adding new genes (knock-in)
- Removing or disrupting genes (knockout)
Purpose of genetically modified organisms
To give organisms new traits, such as disease resistance, faster growth, or the ability to produce valuable substances (like insulin or biofuels).
why is is more difficult to insert genomes into plants and how can it occur
due to their rigid cell wall
1. Electroporation
2. Particle Gun ("Gene Gun")
3. Agrobacterium tumefaciens
Transgenic organisms
Organisms with genes from other species inserted.
Electroporation
Technique using electric field to introduce DNA into cells.
Particle gun
Tiny metal particles coated with DNA are shot into plant cells at high speed.
Agrobacterium tumefaciens
A bacterium that naturally inserts DNA into plant genomes.
Scientists disarm the Ti plasmid (tumor-inducing gene) and replace it with desired genes (like pest resistance or drought tolerance).
Bt toxin gene inserted into crop
make them insect resistant
Recombinant E. coli for Somatotropin
Engineered to produce bovine somatotropin (BST), a hormone that increases milk production in cows.
autotrophic ecoli
Genetically modified to fix carbon dioxide (CO₂) and convert it into sugars.
- Knocking out carbon metabolism genes
- Adding plasmids with pathways for CO₂ fixation
- E. coli becomes capable of producing carbon-based compounds like biodiesel, helping address sustainable fuel production.
genetically engineered vaccines
1) live attenuated vaccines
2) recombinant protein vaccines
3) recombinant vector vaccines
live attenuated viruses
Use a weakened form of the virus/bacteria that still triggers an immune response
recombinant protein vaccines
Use purified proteins from the pathogen (produced in yeast, bacteria, etc.) to stimulate immunity.
recombinant vector vaccines
A harmless virus is used to carry genes from a harmful virus, allowing the body to build immunity without direct exposure to the pathogen.
ethical considerations of biotechnology
- Potential for long-lasting effects in the environment or human health
- Need to balance innovation with safety, transparency, and sustainability
Bacteriophages (t4)
- infect bacteria
- Bind to specific things on the bacteria
- Others have different
- Inject into the bacterioal cell
- Replication to make to particles then releases to get new cells
- Can be lyticor lysogeneic
induction
lysogenic to lytic pathway
types of eukaryotic viruses
enveloped or non enveloped
enveloped virus
has a lipid bilyer that comes from the membrane of the host cell
- Vital proteins in lipid bilyer to bind to eukaryotic cell
- Lipid bilyaer is not as strong as the protein layer of the enveloped then easier to kill
noneveloped
no lipid bilyare but made up of at least 1 or more proteins
Viral genomes
Genetic material of viruses, can be linear or circular.
Virus replication cycle
1. attachment
2. entry and uncoating
3. replication and assembly
4. assembly
5. release
attachment
protein or proteins that attach to one or more celular factors
attachment differences between viruses
Different viruses bind to different host receptors (some only bind to specific organs)
Some bind to different host receptors
Some use only one specific
Some need to bind to two receptor
Some can use multiple receptors
how viruses can enter the cell
1) Virus enters at the cells surface by forming a pore in the cell membrane
2) Envelope virus: that membrane fuses with the cellular membrane to allow the membrane to enter
3) Virus gets endocytose: binds then cellular membrane engulfs and takes in the endosome. Needs to get out of the endosome before becomes lysosome
- Form a pore to eject genome
-Fuse with endosome membrane
- Rip up endosome membrane → now capside can uncoat and reach genome
Replication and protein synthesis differences
Depends on the virus and depends on the location
Endocytosis entry into the cell
binds then cellular membrane engulfs and takes in the endosome. Needs to get out of the endosome before becomes lysosome
- Form a pore to eject genome
- Fuse with endosome membrane
- Rip up endosome membrane → now capsid can uncoat and reach genome
Capsid Uncoating
release of virus from its protective cell and genetic material is released
Replication
Virus genome duplication and protein synthesis.
Assembly
Formation of capsids and genome packaging.
Release Mechanisms
Methods include lysis, budding, and secretion.
Lysis
Cell rupture releasing viral particles.
Budding
When assembly occurs on cell surface → budding → lipid bilyaer comes from cell membrane pinches off and becomes a virus particle
secretion
viral proteins are transported out of the host cell through the cell’s secretory pathway
ways the virus can infect the host cells
1. formation of a proviral state
2. productive
3. chronic
4. latent
Proviral State
Virus integrates into host genome, may cause tumors.
Productive Infection
Continuous virus release via exocytosis.
Chronic Infection
Persistent infection with leftover viral particles.
Latent Infection
Virus present but not actively replicating.
Titer
Concentration of virus per volume unit.
Plaque Assay
Method to quantify viruses using bacterial lawns or eukaryotic lawns
- each white dots is a virus particle
- needs to be diluted
Advantages of plaque assay
- Easy to count
- Can purify virus from each plac
- Dont need as much special machinery
disadvatnages of a plaque assay
Some viruses take 2 weeks
Influenza Virus
RNA virus with segmented genome, enveloped, Orthomyoxiviridae family
Influenza A
Most virulent strain affecting humans. many types
Influenza B
Primarily infects humans and some animals.
- seals and ferrets
Influenza C
Infects humans, dogs, cows, and pigs.
Influenza D
Primarily affects cows.
classifying different influenza viruses
use HA and NA
Hemagglutinin (HA)
Protein for receptor binding in influenza.
- Fushion of viral membrane to the endosome membrane to release segmented genome
Neuraminidase (NA)
Protein mediating viral release.
- controls the virus budding
Sialic Acid Receptors
Key for influenza binding and entry.
influenza life cyle
1. bind to saliac acid recepotrs
2. gets endocytosied
3. HA Fushion of viral membrane to the endosome membrane to release segmented genome
4. Goes into nucleus messenger RNA synthesis
4. Genome not transcribed right away needs to be Synthesized int 5' to 3'
5. RNA replication then er and golgi
6. Assembly is on the cell membrane
7. And the virus buds → NA
studying the flu
1. cell culture
2. animal model
cell culture
- eukaryotic cells in a dish then you infect with a virus then you - look at the immune response
- No t cell and
- Take supernantant proteins → and look to see if new virus have formed
- Take growth curves and compare different influenza virus
mice animal model
morbidity and mortality. Where does that flu virus replicate. Mostly lungs but can go to other parts of the body
Swine Transmission
Involves reassortment and cross-species infection.
- have all the saliac acid receptors so they ca get infected with multiple things at once
Ferret Model
look at their transmission and they have the same saliac acid receptors as humans in nasal and lungs
transmission experiment
- direct contact versus aerosol contact
- Donor ferrets infected with influenza virus
- Contact and aerosol introduedc at 1 dpi
- Nasal washes every 2 days for two weeks
- Nasal washes → determine the titer of the cells