Biochemistry Class 3 - Molecular Biology
1/89
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
90 Terms
What is the monomer for nucleic acids?
Nucleotides and nucleosides
What are nucleotides? Nucleosides?
Nucleotides: sugar, base, phosphate
Nucleoside: sugar and base
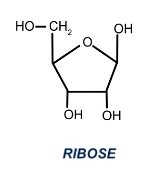
What is ribose? Structure
See image
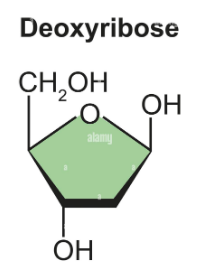
What is deoxyribose? Structure
See image
What are building blocks of DNA?
2’ deoxynucleoside triphosphate
dNTP
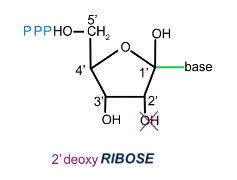
How are dNTPs connected?
3’ O on first dNTP connects to 5’ phosphate of second
Two phosphates leaving second dNTP releases necessary energy
What direction are nucleic acids synthesized? Direction they are written in?
5’ to 3’
How is DNA structured?
5’ to 3’ synthesis
Antiparallel
Complementary bases paired
Bases held together via phosphodiester bond
What are pyrimidines?
Cytosine and Thymine and Uracil
Six membered ring
What are purines?
Adenine and Guanine
Six membered ring stuck to five membered ring
How are A, C, T, G held to their complementary bases?
C and G by three H bonds (more stable)
A and T by two H bonds (less stable)
How do prokaryotic and eukaryotic genomes differ?
Prokaryote: single circular genome, in cytoplasm
Eukaryote: chromosome strings, in nucleus
How do prokaryotes protect themselves from restriction enzymes?
Restriction enzymes: chop up DNA to restrict virus growth
Methylation of DNA prevents REs from binding its own DNA
Supercoil DNA
What does DNA gyrase do?
Supercoils prokaryotic DNA
How is DNA packaged in eukaryotes?
DNA → wrap twice around histone → nucleosome → wind into chromatin
What is a centromere?
Only in eukaryotic genomes
Where spindle fibers attach
Where sister chromatids attach
What is the short arm of a chromosome? Long?
Short: p arm
Long: q arm
What is a telomere?
Only on eukaryotic genomes
Ends of linear chromosomes (made of single and double stranded DNA)
Stabilize end of chromosome by capping it
Consist of short repeats (As and Ts)
What is the central dogma?
DNA → RNA → proteins
Use RNA as an intermediate so you do not damage DNA by working with it directly
What is transcription?
DNA → RNA
Both use nucleotides
What is translation?
RNA → protein
Nucleotides → amino acids
What is the start codon?
AUG
Specify methionine
What are the stop codons?
UAA
UGA
UAG
Do not code for amino acids
How many codons are there?
61
What is the tryptophan codon?
UGG
What are sources of mutations?
Polymerase errors, endogenous damage, exogenous damage, transposons
How do polymerase errors affect DNA?
Cause point mutations, small repeats, or small insertions/deletions
What are point mutations?
Wrong nucleotide put in
What are small repeats?
Polymerase slips off DNA → reputs itself in almost the same place → re-replicates a few bases
What are insertions/deletions?
Insertions: caused by chemicals that trick polymerase into thinking it should put a base somewhere it should not
Deletions: damage causes bases to polymerize in a way that makes them unrecognizable, so polymerase thinks there are none there
What are point mutations? Types and effects
Single base pair change
Missense → changes the amino acid sequence
Nonsense → changes a codon into a stop codon
Silent → no change in amino acid sequence
What are frameshift mutations? Types and effects
Insertions and deletions that change the reading frame
How do exogenous and endogenous damage affect DNA? What causes this?
Endogenous: ROS, physical damage → oxidized DNA, crosslinked bases, physical damage, can lead to polymerase errors
Exogenous: radiation, chemicals → pyrimidine dimers, double strand breaks, translocations, physical damage, intercalation, polymerase errors
What damage do UV and X-ray radiation cause to DNA?
UV: pyrimidine dimers
X-rays: double strand breaks, translocations
What are transposons and how do they affect DNA?
Genes that move around in the DNA
Large insertions/deletions, inversions, duplications
What is the anatomy of a transposon
Transposase enzyme flanked by DNA
Transposase cuts and pastes DNA into other places of DNA
DNA flanking transposons has inverted repeats

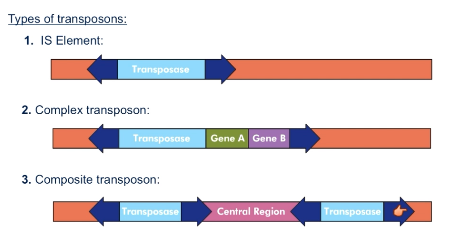
What is an IS element?
Transposase flanked by inverted repeats
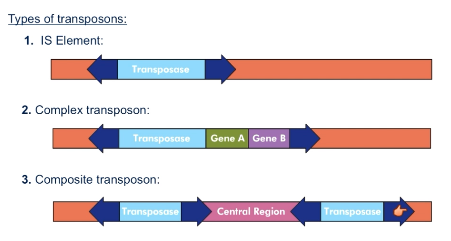
What is a complex transposon?
Transposase and gene(s) flanked by inverted repeats
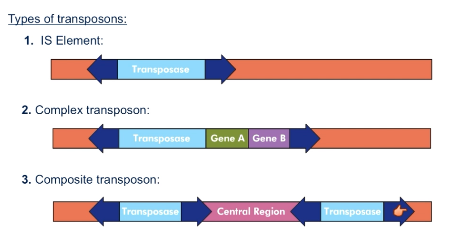
What is a composite transposon?
Two sets of transposons with inverted repeats flanking them
Central region between them
How are transposons inserted?
Transposase is made
Transposase cuts transposon out
Transposase puts the transposon elsewhere
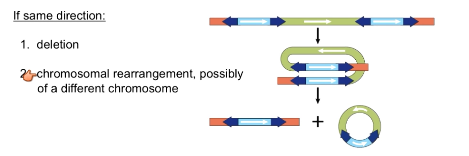
What are effects of composite transposons that run in the same direction?
Deletions
Chromosomal rearrangement (same or other)
What are effects of composite transposons that run in the opposite direction?
Inversions
Chromosomal rearrangement on the same chromosome
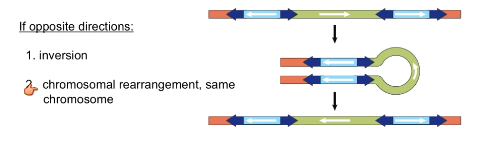
How are polymerase errors repaired?
Mismatch repair pathway (after replication)
Nucleotide excision repair (before replication)
How are transposon errors repaired?
They generally are not
How does mismatch repair work?
Repairs polymerase errors after replication
Recognizes unmethylated DNA as the replicated (messed up) strand
Uses old strand as template
How does nucleotide excision repair work?
Prior to replication
Removes and replaces the bad base
How are endogenous and exogenous DNA damage repaired?
Both: nucleotide excision repair, end joining (homologous and non)
Exogenous only: direct reversal
How does homologous end joining work?
Repairs double stranded DNA breaks
Uses sister chromatid as template
After replication, as it requires sister chromatid
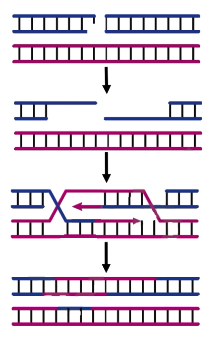
How does nonhomologous end joining work?
Repairs double strand DNA breaks
No sister chromatid for template
Sticks shit together → mutagenic, but better than broken
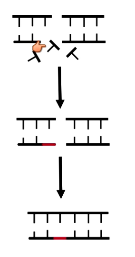
What kinds of errors are caused by nonhomologous end joining?
Translocations
What is direct reversal?
Visible light reverses dimerization caused by UV radiation
What are the rules of DNA replication (4)?
Semiconservative
5’ to 3’ direction
Requires a primer
Requires a template
What does helicase do?
Binds to origin of replication and uncoils DNA
What does topoisomerase do?
Cuts DNA to relax supercoiling caused by helicase
What does primase do?
Puts down the RNA primer
What does DNA polymerase do?
Replicates DNA, proofreads, removes RNA primer
What does ligase do?
Fuses together Okazaki fragments
How is DNA replicated in prokaryotes? AKA Theta replication
1 origin of replication
5 DNA polymerases
What is DNA polymerase III?
In prokaryotes
Main replicating enzyme
5’ to 3’ polymerase and 3’ to 5’ exonuclease activity
Starts 400 bases downstream of the origin
Fast
What is DNA polymerase I?
In prokaryotes
Adds nucleotides at the RNA primer until polymerase III can start
Has 5’ to 3’ polymerase
Has 3’ to 5’ and 5’ to 3’ exonuclease activity
Slow
Helps with DNA excision repair
What does DNA polymerase II do?
5’ to 3’ polymerase and 3’ to 5’ exonuclease
Backup for DNAP III
Does DNA repair
Not necessary for cell to live
What do DNA polymerases IV and V do?
Error prone 5’ to 3’ polymerase activity
DNA repair
Not super important for cell
What do exonucleases do?
Allow errors that were just made to be corrected
Only works at the end of strands, not internally
How does DNA replication in eukaryotes work?
Multiple origins, replication bubbles, several DNA polymerases with complex subunits
What is the end replication problem?
RNA primers add at lagging end, but they can never be put down at the very very end → some cannot be replicated → small single stranded part of DNA (telomere)
Every round of replication shortens the telomere
What is telomerase?
In eukaryotes
Has reverse transcriptase activity and an RNA primer built in
Lengthens telomeres by adding repetitive nucleotide sequences
What is rRNA?
ribosomal RNA
Catalytic part of ribosome
What is mRNA?
Messenger RNA
Sequence of codons determines the amino acid sequence of the protein
What is tRNA?
Transfer RNA
Carries amino acids to ribosomes
What is hnRNA?
Heterogeneous nuclear RNA
Initial unprocessed transcript
What are siRNA and miRNA?
Micro RNA and Small Interfering RNA
Help regulate gene expression
How are replication and transcription similar/different?
Both: start site, 5’ to 3’, DNA template
Replication: no stop site, needs primers, proofreading
Transcription: stop site, no primer, no proofreading
What are template vs coding strands?
Coding: AKA sense, same sequence as RNA
Template: AKA antisense, used to make RNA
How does transcription work?
RNAP binds to promoter → reaches start site → starts transcribing → reaches stop site → polyadenylase adds poly A tail
How is translation primarily regulated? Two types
At the transcription level (once you have mRNA, you basically have proteins)
Via promoters or DNA binding proteins
How do promoters regulate translation?
Strong promoters have high affinity for RNAP → lots of transcription
Weak ones have low affinity for RNAP → less transcription
How do DNA binding proteins regulate translation?
Repressors: bind DNA and prevent transcription
Enhancers: bind activators → bind transcription factors in proximity to promoter → increase transcription
How are prokaryotic and eukaryotic transcription different?
Prokaryotic: transcription and translation at same place/time (cytosol); no mRNA processing; polycistronic; one RNAP
Eukaryotic: transcription in nucleus, translation in cytosol; mRNA must be processed; monocistronic; three RNAPs
How is mRNA processed?
In eukaryotes
5’G cap added
3’ poly A tail
Splicing to remove introns
What does it mean to be polycistronic?
From one mRNA, many different proteins can be made
What do RNAP I, II, and III do?
In eukaryotes
RNAP I: transcribes rRNA
RNAP II: transcribes mRNA
RNAP III: transcribes tRNA
What is the anatomy of a tRNA?
Amino acid attaches at 3’ end
Anticodon at second loop
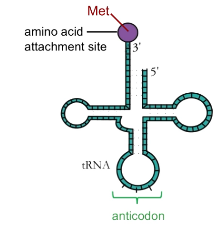
What is wobble pairing?
Third position anticodon has more flexible base pairing than the others
Reduces how many codons you need to make
G in tRNA can bind to U or C on mRNA
U in tRNA can bind to A or G in mRNA
A always binds U
C always binds G
What is inosine?
Adenine tRNA can convert into inosine
It can bind to A, U, or C
How are tRNAs made?
Aminoacyl tRNA synthetase loads amino acid on the tRNA
Costs 2 ATP per amino acid
How do prokaryotic and eukaryotic ribosomes differ?
Prokaryotic: 50S large subunit, 20S small subunit; total 70S
Eukaryotic: 60S large subunit, 40S small subunit; total 80S
S is a measure of how fast you sink in a centrifuge (aka size)
How do ribosomes work?
Growing protein held on P site
Amino acids added at the A site
mRNA binds to ribosome large subunit (first codon in P site, second in A) → tRNA binds to P-site → tRNA binds to A site → break bond between tRNA and amino acid at P site (release 2 ATP) → translocation down one codon → → reach stop codon (no tRNA for this) → bind release factor → breaks final tRNA - amino acid bond
ATP released is used to make peptide bonds
How much ATP is used at each step of making a protein?
tRNA loading: 2 per amino acid
Initiation: 1 per peptide
A site binding: 1 (per amino acid minus 1)
Translocation: 1 (per amino acid minus 1)
Termination: 1 per peptide
ATP used = # of amino acids x 4
How are proteins post-translationally modified?
Folded by chaperones
Can add covalent modifications (disulfide bridges, glycosylation, phosphorylation)
Processing (ex: removal of some parts to activate it)
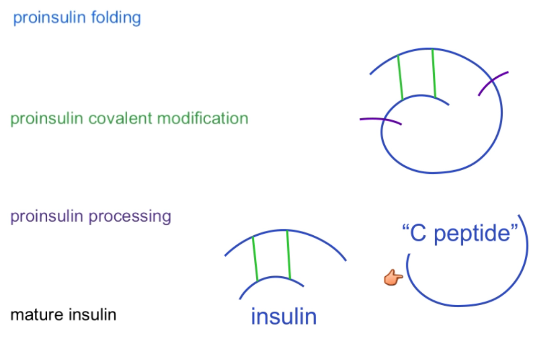
How is insulin post-translationally processed?
Folded → disulfide bridges → cleavage into insulin and C peptide (non functional but can be measured)