last half
5.0(1)
Card Sorting
1/46
There's no tags or description
Looks like no tags are added yet.
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
1
New cards
virus
genetic element that can multiply only inside a living cell called the host cell
2
New cards
obligate intracellular parasites
viruses rely on the host cell for energy, metabolic intermediates, and protein synthesis
3
New cards
Nature of Viruses
Viruses are not considered living entities
* **not included on the tree of life**, but do infect cells in all three domains (Archaea, Bacteria, Eukarya)
* **are not cells** though their genomes encode those functions needed to multiply and have structurally intricate extracellular form called the **virion**
* **Cannot reproduce** unless the virion itself, or in some cases its genome only, has gained entry into a suitable growing host cell, a process called **infection**
* **not included on the tree of life**, but do infect cells in all three domains (Archaea, Bacteria, Eukarya)
* **are not cells** though their genomes encode those functions needed to multiply and have structurally intricate extracellular form called the **virion**
* **Cannot reproduce** unless the virion itself, or in some cases its genome only, has gained entry into a suitable growing host cell, a process called **infection**
4
New cards
infection
Cannot reproduce unless the virion itself, or in some cases its genome only, has gained entry into a suitable growing host cell
5
New cards
structure of the virion
* **nucleocapsid**
* capsid proteins - composed of capsomeres
* nucleic acid ( virus genome)
* naked/**envelope**
* an outer layer, most often composed of a phospholipid bilayer taken from the host cell membrane
* capsid proteins - composed of capsomeres
* nucleic acid ( virus genome)
* naked/**envelope**
* an outer layer, most often composed of a phospholipid bilayer taken from the host cell membrane
6
New cards
capsid
composed of a number of individual protein molecules called **capsomeres** that are often arranged in a precise and highly repetitive pattern around the nucleic acid
7
New cards
function of virions
* Virion protects the viral genome when the virus is outside the host cell
* proteins on the virion surface are important in attaching it to its host cell - **adsoprtion**
* proteins on the virion surface are important in attaching it to its host cell - **adsoprtion**
8
New cards
Virion symmetry
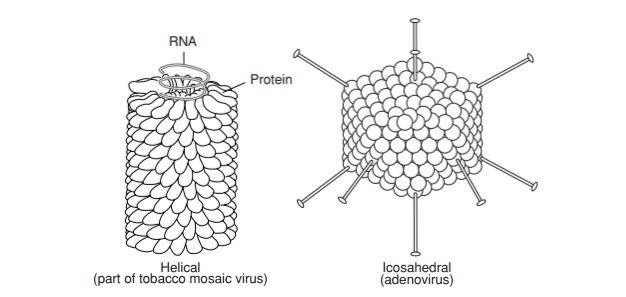
1. **helical symmetry** - rod shaped
1. Ebola virus
2. **icosahedral symmetry** - spherical
1. Human papillomavirus virion
9
New cards
Purpose of viruses
a virus’ main purpose is to replicate, therefore , it must induce a living host cell to synthesize all the essential components needed to make new virions
* because of all the biosynthetic and energy required, viruses cannot replicate in dead host cells
* because of all the biosynthetic and energy required, viruses cannot replicate in dead host cells
10
New cards
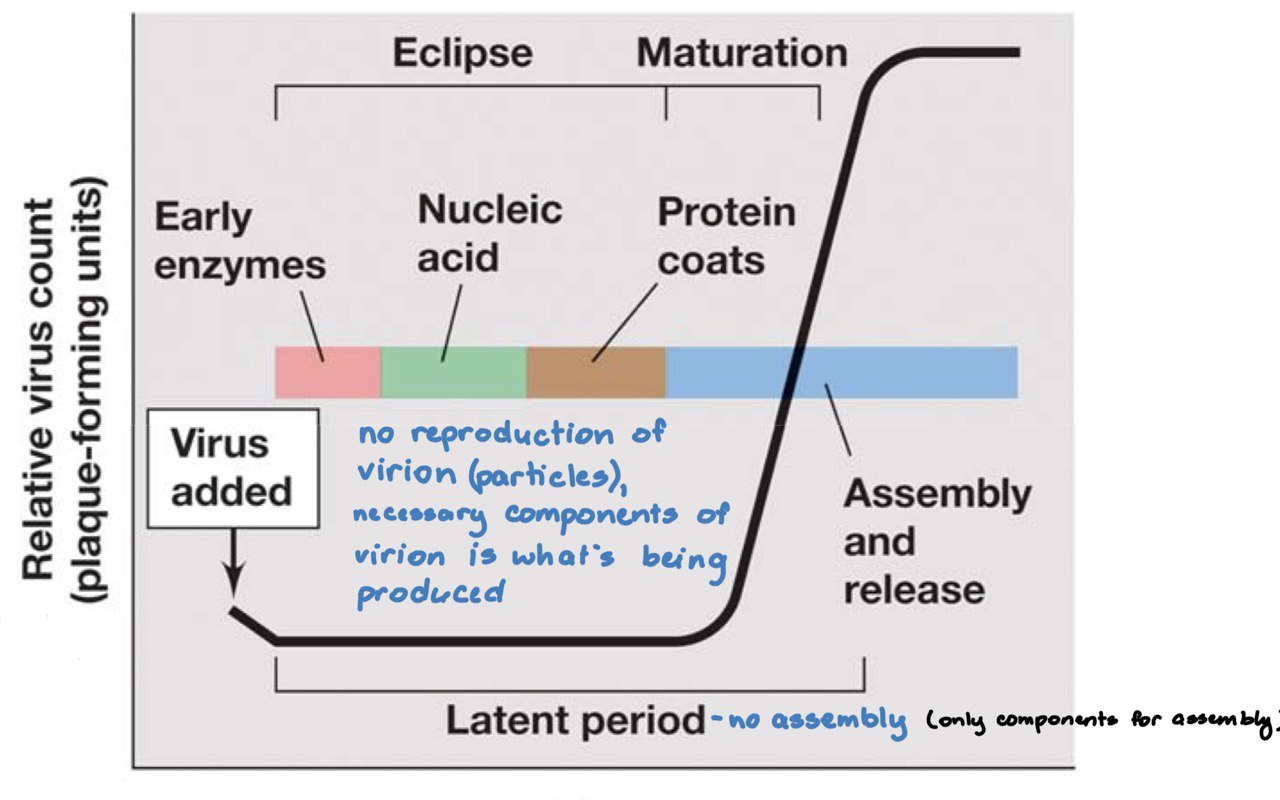
Viral Replication Cycle
1. **adsorption** of phage virion
2. **penetration** of nucleic acid
3. **biosynthesis** - takes over the host cell, using its resources and mechanisms to make more viral genomes and viral proteins
4. **assembly** (and packaging of new viruses)
5. **cell lysis** - release of new virions, need new hosts to provide more resources for new virions
\
**Difference:** nucleic acid is inserted in prokaryotic cells and leaves the capsid behind, while the whole virion is taken up by the animal and plant cells (eukaryotes)
11
New cards
Protein synthesis categories
1. **Early proteins** - enzymes such as nucleic acid polymerases for **genome replication** and other factors used to shut down the host’s transcription and translation
2. **late proteins** - structural components and other components needed for packaging and assembly
\
**bacterial virus:** 20-60 mins
**animal virus:** 8-40 hours
12
New cards
Early proteins
enzymes such as nucleic acid polymerases for **genome replication** and other factors used to shut down the host’s transcription and translation
* don’t want the host cells to make their own proteins, therefore you shut it down to make cells make virus-specific proteins instead
* don’t want the host cells to make their own proteins, therefore you shut it down to make cells make virus-specific proteins instead
13
New cards
Burst size
refers to average number of virions released
14
New cards
Late proteins
structural components and other components needed for packaging and assemby
15
New cards
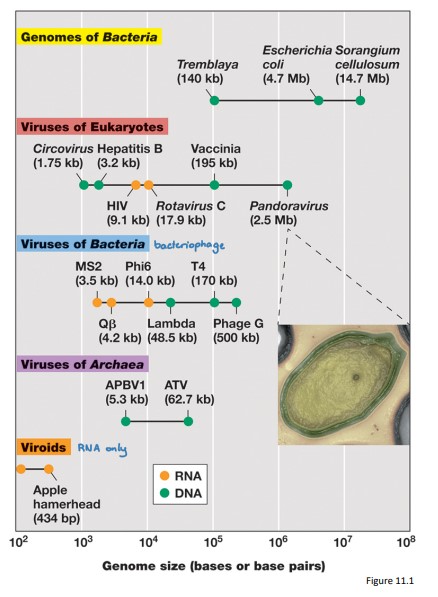
Range in Virus Sizes
Viral genomes vary almost a thousand‐fold in size from smallest to largest and are grouped by genome structure (dsDNA, ssDNA, dsRNA, ssRNA)
16
New cards
Baltimore Classification of Viral Genomes
* a chart based on the relationship of the viral genome to its mRNA
* **7 classes of viruses**:
* 3 have DNA genomes and
* 4 have RNA genomes
* **7 classes of viruses**:
* 3 have DNA genomes and
* 4 have RNA genomes
17
New cards
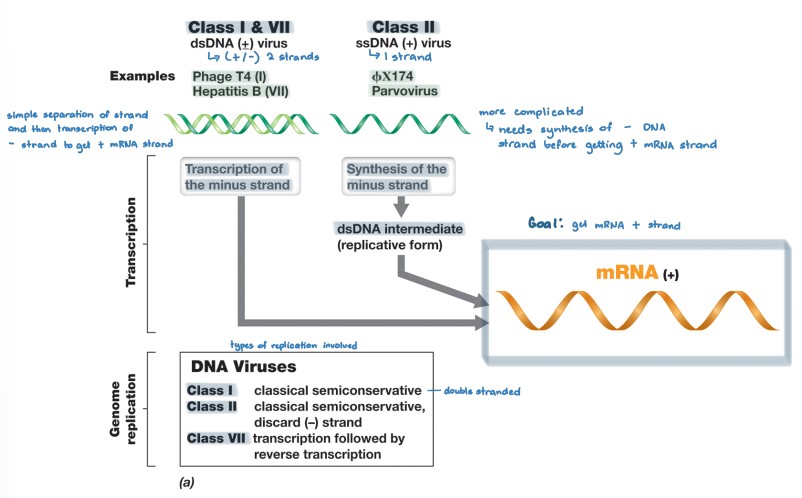
DNA viruses
* class I (+/-)
* uses the same mechanism as the host to replicate and produce mRNA production and genome replication
* class II (+)
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* dsDNA intermediate form used as new genome copies
* class VII (+/-)
* use reverse transcriptase to replicate from (-) mRNA to (+) DNA
* uses the same mechanism as the host to replicate and produce mRNA production and genome replication
* class II (+)
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* dsDNA intermediate form used as new genome copies
* class VII (+/-)
* use reverse transcriptase to replicate from (-) mRNA to (+) DNA
18
New cards
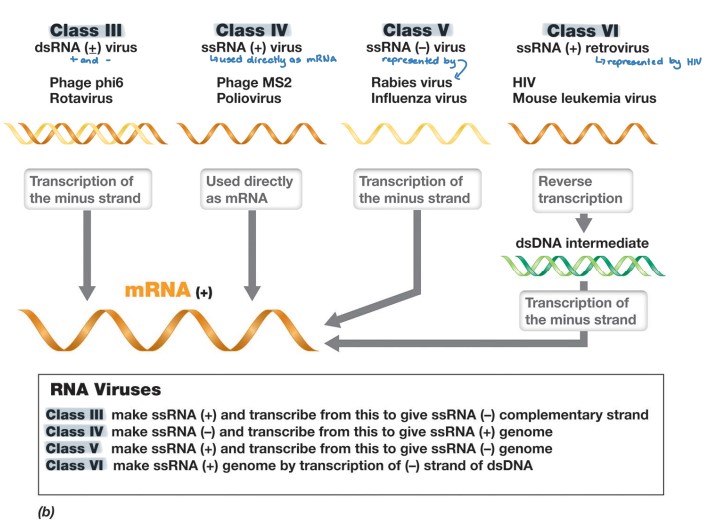
RNA viruses
* Class III (+/-)
* (-) RNA strand → (+) mRNA
* Class IV (+)
* genome is used directly as + mRNA
* Class V (-)
* (-) RNA strand → (+) mRNA via RNA replicase
* Class VI (+)
* uses reverse transcriptase of dsDNA intermediate to replicate
* (+) RNA → dsDNA intermediate → (-) DNA → (+) mRNA
* (-) RNA strand → (+) mRNA
* Class IV (+)
* genome is used directly as + mRNA
* Class V (-)
* (-) RNA strand → (+) mRNA via RNA replicase
* Class VI (+)
* uses reverse transcriptase of dsDNA intermediate to replicate
* (+) RNA → dsDNA intermediate → (-) DNA → (+) mRNA
19
New cards
bacteriophages/phages
Bacterial viruses
* Intensively studied as model systems for the molecular biology and genetics of virus replication (particularly T4)
* Intensively studied as model systems for the molecular biology and genetics of virus replication (particularly T4)
20
New cards
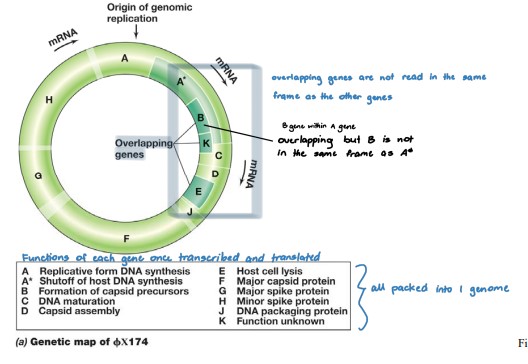
Phage ϕX174
* Class II DNA - ssDNA (+)
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* Icosahedral (sphere), \~25 nm in diameter, circular genome of 5386 nucleotides
* Infection process
* Binds specifically to LPS surface on E. coli
* genome is inserted, leaving the capsid outside
* DNA replication via **rolling circle replication**
* Virion assembly
* SSBs (proteins that regulates processes such as transcription and translation) removed as ssDNA is packaged into the capsid
* Protein E inhibits peptidoglycan synthesis, cell walls start having holes, promotes cell lysis
* burst size = 500 virions
\
**Genome has overlapping genes**
* insufficient DNA to encode all viral‐specific proteins
* parts of the genome are transcribed in more than one reading frame
* Genes reside within genes but with different promoters and frame reads • For gene A, same mRNA can be translated into 2 different proteins (A and A\*) each with a different start codon
\
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* Icosahedral (sphere), \~25 nm in diameter, circular genome of 5386 nucleotides
* Infection process
* Binds specifically to LPS surface on E. coli
* genome is inserted, leaving the capsid outside
* DNA replication via **rolling circle replication**
* Virion assembly
* SSBs (proteins that regulates processes such as transcription and translation) removed as ssDNA is packaged into the capsid
* Protein E inhibits peptidoglycan synthesis, cell walls start having holes, promotes cell lysis
* burst size = 500 virions
\
**Genome has overlapping genes**
* insufficient DNA to encode all viral‐specific proteins
* parts of the genome are transcribed in more than one reading frame
* Genes reside within genes but with different promoters and frame reads • For gene A, same mRNA can be translated into 2 different proteins (A and A\*) each with a different start codon
\
21
New cards
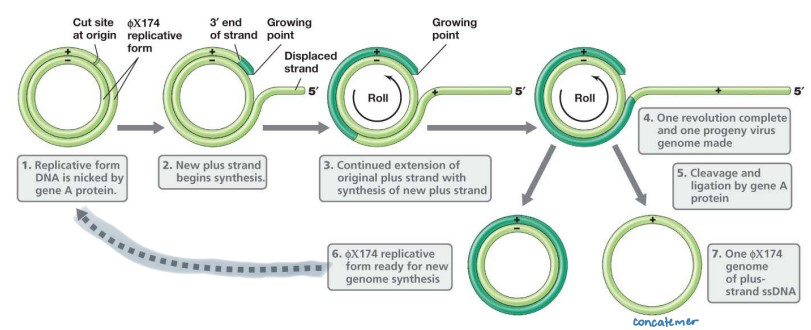
Phage ϕX174 Replication (ssDNA +)
**Rolling Circle Replication**
* Uses hosts enzymes to make replicative form
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* transcribe and translate until increased levels of protein A
* Viral A protein cuts at specific + strand of circular dsDNA
* 5′ end is displaced
* 3′ end serves as a primer for DNA synthesis
* (+) strand synthesis is initiated
* dNTPs added to 3′ end, using exposed (‐) strand as template
* 5′ end of the (+) strand peels away exposing more template
* SSB’s (ssDNA binding phage proteins) coats the displaced 5′ end to prevent positive strand from serving as a template for DNA polymerase
* Continued rotation of circle through replicating site results in complete linear copy of (+) strand aka **concatemers**
* Viral A protein cuts (+) ssDNA, ligates ends to make circular (+) ssDNA genome
* SSBs removed as ssDNA is packaged into the capsid
* (+/‐) dsDNA replicative form ready for another round of asymmetric replication
* Uses hosts enzymes to make replicative form
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* transcribe and translate until increased levels of protein A
* Viral A protein cuts at specific + strand of circular dsDNA
* 5′ end is displaced
* 3′ end serves as a primer for DNA synthesis
* (+) strand synthesis is initiated
* dNTPs added to 3′ end, using exposed (‐) strand as template
* 5′ end of the (+) strand peels away exposing more template
* SSB’s (ssDNA binding phage proteins) coats the displaced 5′ end to prevent positive strand from serving as a template for DNA polymerase
* Continued rotation of circle through replicating site results in complete linear copy of (+) strand aka **concatemers**
* Viral A protein cuts (+) ssDNA, ligates ends to make circular (+) ssDNA genome
* SSBs removed as ssDNA is packaged into the capsid
* (+/‐) dsDNA replicative form ready for another round of asymmetric replication
22
New cards
concatemers
original copy of ssDNA strand
23
New cards
Phage M13
* Class II DNA - ssDNA (+)
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* Filamentous virus (long and thin) with helical symmetry, circular genome
* **attaches to the pilus** of its host cell and leaves capsid outside of cell
* uses **rolling circle replication** but does not undergo cell lysis when virions are released
* non-lytic release of virions, therefore the integrity of the host cell is maintained
* facilitated by covering M13 DNA with **coat proteins** as it exits from the cell envelope
* Mature M13 virions **do not accumulate** in the cell as with typical lytic bacteriophages
* **chronic infection (steady state infection)**: Infected cells continue to grow, and typical viral plaques are not observed - long term, lesser effect
* 1,000 virions released per generation
* (+) ssDNA → dsDNA intermediate form → (+) mRNA
* Filamentous virus (long and thin) with helical symmetry, circular genome
* **attaches to the pilus** of its host cell and leaves capsid outside of cell
* uses **rolling circle replication** but does not undergo cell lysis when virions are released
* non-lytic release of virions, therefore the integrity of the host cell is maintained
* facilitated by covering M13 DNA with **coat proteins** as it exits from the cell envelope
* Mature M13 virions **do not accumulate** in the cell as with typical lytic bacteriophages
* **chronic infection (steady state infection)**: Infected cells continue to grow, and typical viral plaques are not observed - long term, lesser effect
* 1,000 virions released per generation
24
New cards
Budding
* The viral DNA crosses the cell envelope through a **channel constructed from virus‐encoded proteins**
* Viral capsid proteins are inserted into the host cell’s membrane to form patches (aggregates of proteins)
* As this occurs, the DNA is coated with phage proteins that have been embedded in the cytoplasmic membrane
* A process also known as **blebbing** (non-lytic cycle)
\
Process
1. **Infection:** phage attaches to the host’s pilus and inserts the DNA, leaving the capsid behind
2. **Biosynthesis:** replicates using rolling circle replication, transcription and translation increases pV, which stops replication and initiates assembly
3. pV coats the ssDNA and the capsid protein in the host’s cell membrane take up DNA and start to assemble
4. As more genomes enter the cell membrane, p8 replaces p5 and starts the **packaging** process
5. p3 and p6 attach to the phage to form coat and pIII releases particle from inner membrane allowing the phage to be exported
6. p4 is secreted allowing **budding** to occur
* Viral capsid proteins are inserted into the host cell’s membrane to form patches (aggregates of proteins)
* As this occurs, the DNA is coated with phage proteins that have been embedded in the cytoplasmic membrane
* A process also known as **blebbing** (non-lytic cycle)
\
Process
1. **Infection:** phage attaches to the host’s pilus and inserts the DNA, leaving the capsid behind
2. **Biosynthesis:** replicates using rolling circle replication, transcription and translation increases pV, which stops replication and initiates assembly
3. pV coats the ssDNA and the capsid protein in the host’s cell membrane take up DNA and start to assemble
4. As more genomes enter the cell membrane, p8 replaces p5 and starts the **packaging** process
5. p3 and p6 attach to the phage to form coat and pIII releases particle from inner membrane allowing the phage to be exported
6. p4 is secreted allowing **budding** to occur
25
New cards
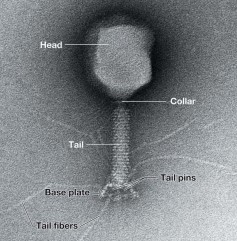
Phage T4
* class I (+/-) dsDNA
* (-) DNA → (+) mRNA
* large, icosahedral tail with helical tail surrounded by a contractile sheath, 170 kbp folded **linear genome**
* Tail structure
* tail ends at base plate
* base plate consists of tail fibers and pins
* virulent - actively reproduce inside the host cell
* lytic - reproduction leads to lysis of the host cell
* uses similar replication mechanism as host cell - “primer” problem
* (-) DNA → (+) mRNA
* large, icosahedral tail with helical tail surrounded by a contractile sheath, 170 kbp folded **linear genome**
* Tail structure
* tail ends at base plate
* base plate consists of tail fibers and pins
* virulent - actively reproduce inside the host cell
* lytic - reproduction leads to lysis of the host cell
* uses similar replication mechanism as host cell - “primer” problem
26
New cards
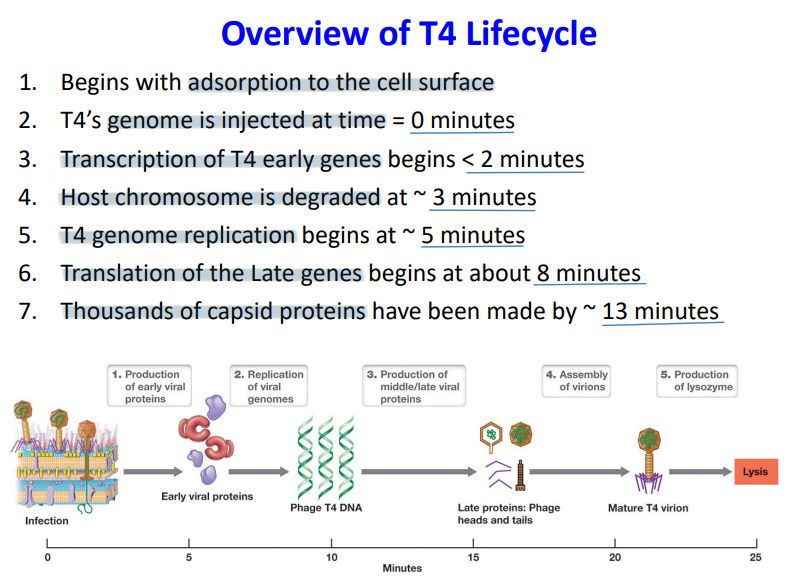
T4 Phage Infection
1. Adsorption - tail fibers contacts LPS molecule (phage receptor) of E.coli
2. Attachment - baseplate and tail pins contact surface of outer membrane
3. Penetration - tail sheath contracts and injects genome by using T4 lysozyme that degrades cell surface
1. degradation products cause sheath protein contraction that uses atp to hydrolyze 24 rings into 12 rings
27
New cards
T4 Phage Transcription and Translation
1. after 1 minute of the phage entering the cytoplasm, the host’s DNA and RNA synthesis stop
2. transcription of phage-specific genes begin
1. **early genes**
1. **T4 nuclease** that stops host gene expression by degrading E. coli chromosome and provides nucleotides for replication of phage genome
2. early gene **promoters** that are recognized by E.coli σ70/RNA polymerase holoenzyme
3. **enzymes** for synthesis and glucosylation of unusual T4 base 5‐ hydroxymethylcytosine (HMC)
1. by adding sugar and “tagging” it, HMC is protected from T4 nuclease and the host’s endonuclease
4. **T4** **replisome** - uses its own replisome because it can’t rely on the hosts to replicate (begins 5 mins after infection and continues for about 20 mins)
5. **Proteins** that modify host RNA polymerase - T4 does not have its own RNA polymerase therefore it modifies the host’s to recognize the phage promoters of middle genes
2. **middle genes and late genes**
1. requires more proteins to modify the host’s RNA polymerase to recognize late gene promoters because late genes become even more phage specific ( T4‐encoded sigma factor)
2. **structural proteins** (capsid, tail, sheath)
3. **packaging motor proteins** - inserting genes into the head
4. **T4 lysozyme** for degradation of peptidoglycan and eventual lysis of cell
\
**Early genes:** E. coli σ70/RNA polymerase holoenzyme
**Middle genes:** phage protein packed onto E. coli σ70/RNA polymerase holoenzyme
**Late genes:** very specific T4‐encoded sigma factor (group of genes that encode for assembly pieces)
28
New cards
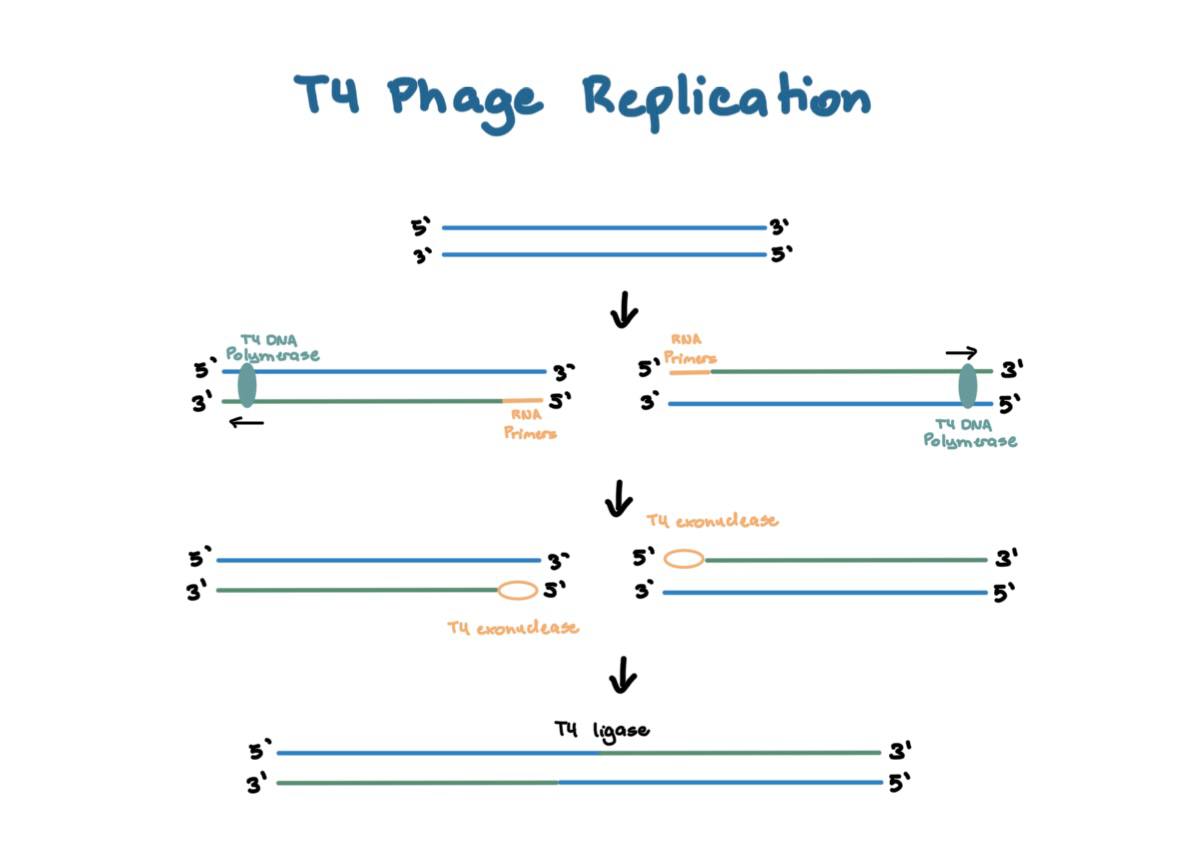
T4 Phage Replication
1. dsDNA unwinds and splits into 2
2. **RNA primers** are added by T4 Primase on the 5’ ends
3. Replication by **T4 DNA polymerase** which results in a linear genome package
4. Primer is degraded by **T4 exonuclease** which results in a 5’ gap (primer problem)
5. **T4 ligase** combines the complementary overhangs and form **concatemers**
29
New cards
T4 Phage Assembly
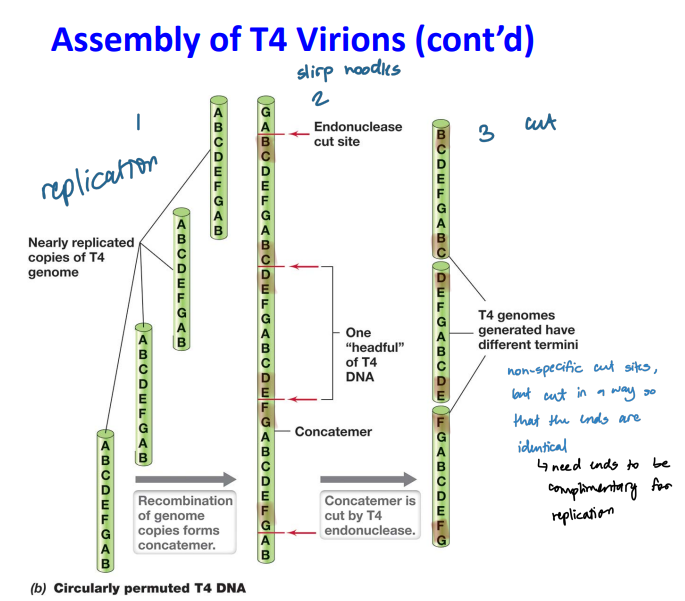
1. **T4 endonuclease** cut the long genome after replication at no specific sequence
1. **headful packaging**
2. The linear segment has a full T4 genome plus a little extra
3. **Circular permutation:** genomes with the same set of genes but arranged in a different order
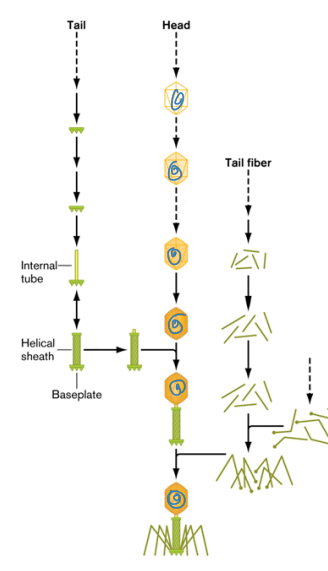
2. **Baseplate and tube (tail) assembly**
1. Baseplate proteins are assembled
2. Tail pins added onto the baseplate
3. Helical tube is added onto the baseplate
4. Sheath proteins added around tube
3. **Capsid assembly**
1. capsids form a prohead
2. one end of the concatemer is drawn into the prohead until the prohead is full and the concatemer is cut to fill the next prohead (sucking noodles)
1. Genome is pumped into prohead by a ATP‐driven packaging motor
4. Tail fiber assembly
1. Tail proteins assembled to form tail fibers
2. Tail fibers added to mature tail as last step before maturation
30
New cards
T4 Life cycle
\
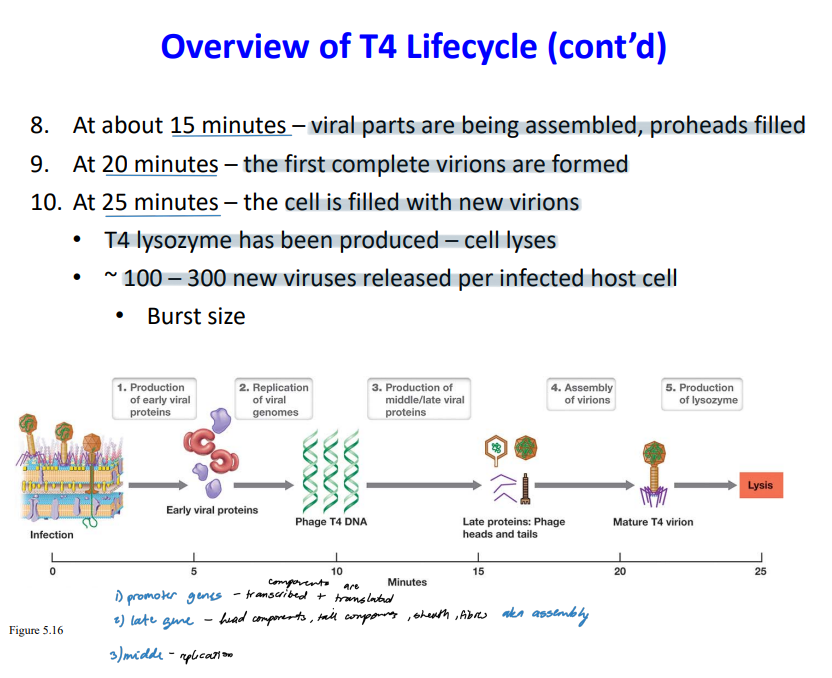
31
New cards
T7 Phage
* dsDNA (±)
* smaller than T4, dsDNA (±)
* virulent and lytic
* encodes for its own T7 RNA polymerase which recognizes only T7 gene promoters
* linear genome - packaging is achieved by very specific cutting sites by T7 endonuclease
* smaller than T4, dsDNA (±)
* virulent and lytic
* encodes for its own T7 RNA polymerase which recognizes only T7 gene promoters
* linear genome - packaging is achieved by very specific cutting sites by T7 endonuclease
32
New cards
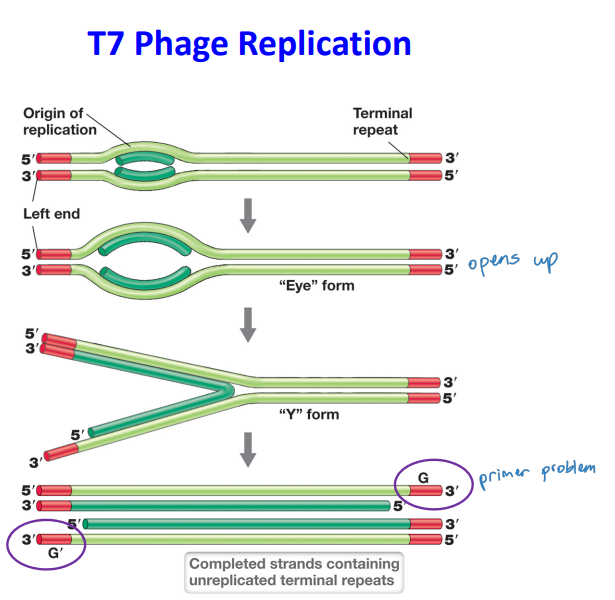
T7 Phage Replication
Replicates linear genome in a manner different from T4 phage
* unreplicated terminal repeats are paired via DNA polymerase and ligase activity to form a concatemer
* Packaging of genomes is achieved by specific cutting by T7 endonuclease
* single stranded cuts are made at specific sites, dna polymerase completes the single strand
* results in terminal repeats but not identical ends
* unreplicated terminal repeats are paired via DNA polymerase and ligase activity to form a concatemer
* Packaging of genomes is achieved by specific cutting by T7 endonuclease
* single stranded cuts are made at specific sites, dna polymerase completes the single strand
* results in terminal repeats but not identical ends
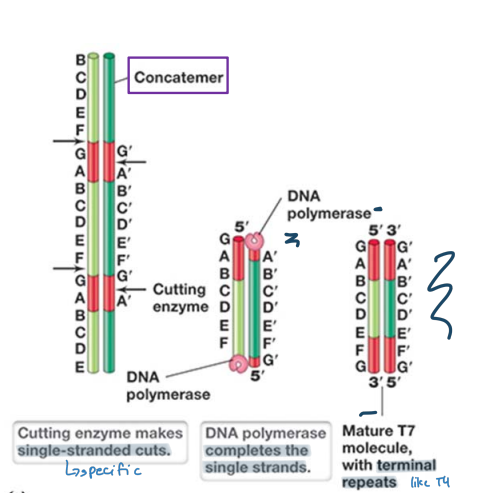
33
New cards
Lysogeny
* temperate phage integrates into the host chromosome → **lysogenic state**
* virus is now known as **prophage**
* few genes are transcribed, just enough to stay in the **dormant state**
* Virus genome is replicated in synchrony with host chromosome and passed to daughter cells at cell division - **vertical transmission**
* Integrated viral genome may confer genetic properties on the bacterial host cell → **lysogenic conversion**
* A bacterial cell that harbors a temperate virus is called a **lysogen**
* virus is now known as **prophage**
* few genes are transcribed, just enough to stay in the **dormant state**
* Virus genome is replicated in synchrony with host chromosome and passed to daughter cells at cell division - **vertical transmission**
* Integrated viral genome may confer genetic properties on the bacterial host cell → **lysogenic conversion**
* A bacterial cell that harbors a temperate virus is called a **lysogen**
34
New cards
Lytic
* Occurs immediately upon infection of host cell or upon excision of the temperate phage from the host chromosome
* Viral genome is replicated, transcribed and translated
* Synthesis of new viral particles
* Host cell lysis releasing virions
* Viral genome is replicated, transcribed and translated
* Synthesis of new viral particles
* Host cell lysis releasing virions
35
New cards
Lytic or Lysogenic
* Generally, the decision for a temperate phage to go through the lytic or lysogeny pathway is based on **environmental conditions**.
* lysogenic: high MOI (lots of people) + low nutrients (empty fridge) = dormant
* lytic: low MOI (one person) + high nutrients (full fridge) = lytic
* Cell stress conditions can “induce” the excision of the prophage and proceed through the lytic pathway
\
* lysogenic: high MOI (lots of people) + low nutrients (empty fridge) = dormant
* lytic: low MOI (one person) + high nutrients (full fridge) = lytic
* Cell stress conditions can “induce” the excision of the prophage and proceed through the lytic pathway
\
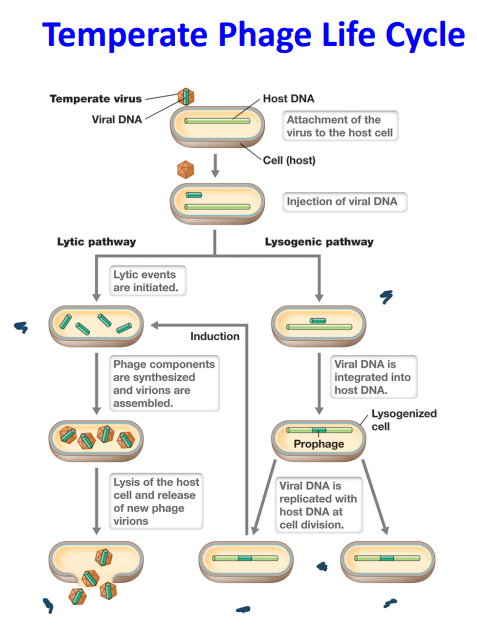
36
New cards
λ phage
* linear dsDNA
* icosahedral head with a long non‐contractile tail
* linear genome in phage’s head but circularizes once it enters E. coli cell
* does not have redundant ends like T4 and T7
* Tail tip binds to E. coli’s LamB protein, which is the outer membrane porin involved in maltose transport
* icosahedral head with a long non‐contractile tail
* linear genome in phage’s head but circularizes once it enters E. coli cell
* does not have redundant ends like T4 and T7
* Tail tip binds to E. coli’s LamB protein, which is the outer membrane porin involved in maltose transport
37
New cards
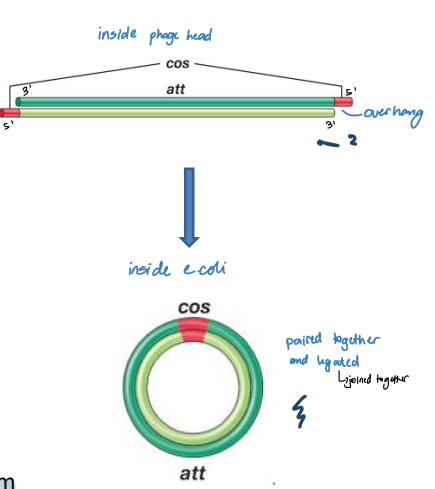
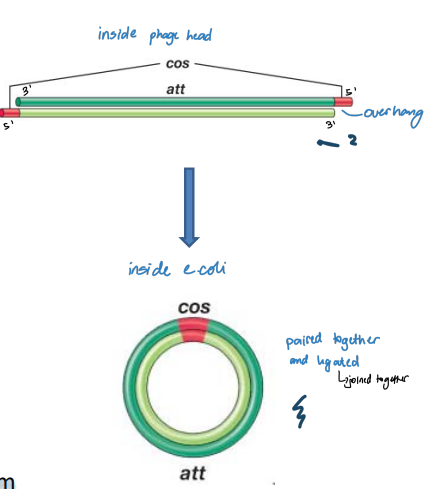
Circularization of λ Genome
Phage head:
* A 12 nucleotide segment at 5′ end of each strand is single-stranded and unpaired (cos sites)
* Sequences in ss regions are complementary to each other
* Can form base pairs
* Cohesive (sticky) ends
\
In E. coli cell:
* Circularization of the genome
* Base pairing between the cos sites
* Host DNA ligase seals nicks to form circular ds replicative form
* A 12 nucleotide segment at 5′ end of each strand is single-stranded and unpaired (cos sites)
* Sequences in ss regions are complementary to each other
* Can form base pairs
* Cohesive (sticky) ends
\
In E. coli cell:
* Circularization of the genome
* Base pairing between the cos sites
* Host DNA ligase seals nicks to form circular ds replicative form
38
New cards
Integration of λ phage = (prophage) Lysogeny
Integration depends on the specific sequence in the λ genome
* Designated the **attP site,** almost opposite cos sites, **base pairs with E. coli’s attB site**, which is a homologous sequence that exists on E. coli’s chromosome (between galactose operon and biotin operon)
* Phage-encoded integrase promotes strand exchange (i.e. recombination) by cleaving phage and chromosomal strands which are then ligated together
* Order of λ genes has been changed – **permuted**
* Designated the **attP site,** almost opposite cos sites, **base pairs with E. coli’s attB site**, which is a homologous sequence that exists on E. coli’s chromosome (between galactose operon and biotin operon)
* Phage-encoded integrase promotes strand exchange (i.e. recombination) by cleaving phage and chromosomal strands which are then ligated together
* Order of λ genes has been changed – **permuted**
39
New cards
Lysogen Unusual Properties
1. Immune to **superinfection** of the same phage (due to the CI repressor)
2. Spontaneous induction of the prophage **(lytic cycle)** - some lyse and release a normal burst size of phages
3. Cell stress conditions can induce prophage to enter the lytic cycle and lyse cell (don’t want to die, therefore go lytic to exit the phage)
4. prophage may be spontaneously lost at low frequency - curing
40
New cards
λ Phage Genome
Three classes of genes:
1. Immediate early
2. Immediate delayed
1. Late
1. Immediate early
2. Immediate delayed
1. Late
41
New cards
Lysogenic Conversion
Prophage induces a phenotypic change in the host cell
1. Changes to cell surface structure
2. Toxin production
1. Changes to cell surface structure
2. Toxin production
42
New cards
Gene Transfer
**Horizontal gene transfer:** movement of genes between cells that are not direct descendants of another
**Vertical gene transfer:** movement of genes between cells that are direct descendants of another (mother to daughter cells)
**Vertical gene transfer:** movement of genes between cells that are direct descendants of another (mother to daughter cells)
43
New cards
Competence
* Physiological state of bacterial cells that are able to take up exogenous DNA
* A cell that is able to take up DNA and be transformed is said to be competent
* A cell that is able to take up DNA and be transformed is said to be competent
44
New cards
Competence is directly linked to pili
* Mechanisms exist to account for differences in cell envelope structure for gram negative and gram positive bacteria
* pili Binds and facilitates uptake of DNA into cell by retraction
* pili Binds and facilitates uptake of DNA into cell by retraction
45
New cards
resolvase enzyme
an enzyme that cleaved on a horizontal or vertical plane
\
1. **Patches (horizontal)** – no recombination (no exchange of markers) with short heteroduplex regions
2. **Splices (vertical)** – recombination has occurred (exchange of markers) with short heteroduplex regions
Heteroduplex regions are mismatched regions which resolved by DNA repair or by DNA replication (2 molecules each with slightly different sequence)
\
1. **Patches (horizontal)** – no recombination (no exchange of markers) with short heteroduplex regions
2. **Splices (vertical)** – recombination has occurred (exchange of markers) with short heteroduplex regions
Heteroduplex regions are mismatched regions which resolved by DNA repair or by DNA replication (2 molecules each with slightly different sequence)
46
New cards
Episome
plasmid that can integrate itself into host chromosome
47
New cards