Grapevine genomics
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
How many sets of chromosomes does a grapevine have? And how many chromosomes do they have?
Two copies of each chromosome so they are diploids
19 chromosome pairs (2N = 38)
Why do we care about grapevine genomics?
As they influence bunch size, architecture, colour, flavour, leaf shape, growing habit, interaction with environment etc
What limitations are involved with short-read genome sequencing?
Short reads (300-500 DNA base pairs long) result in little overlap meaning there are more gaps in the draft genome and it is hard to assemble. Missing sequence leads to missed genes and limits biological interpretation
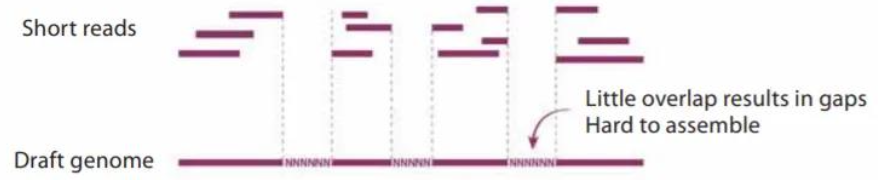
What are the benefits of long-red sequencing for genome assembly?
Long-read sequencing helps to obtain unfragmented genome assemblies
Long-reads (>1000 DNA base pairs up to 2 million base pairs long) result in overlapping reads with no gaps, meaning they are easy to assemble, and the draft genome is complete. Giving a comprehensive structural, functional and organised picture of the genome
Long-reads allows us to independently assemble both copies of a diploid genome
If we can’t assemble the two copies of the genome, we are blind to the genetic diversity of each cultivar
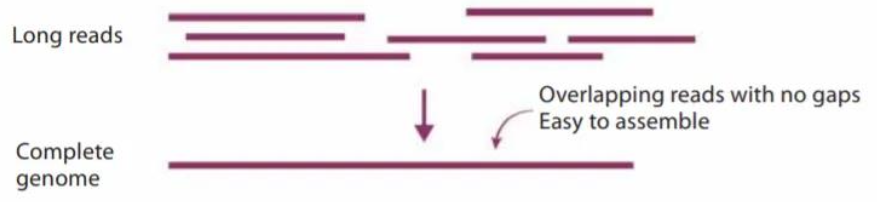
Why is assembling grapevine genomes challenging?
Assembling grapevine genomes is challenging due to high heterozygosity, cell layer mutations and high abundance of repetitive elements:
grapevine cultivars are F1 crosses between distantly related parents
grapevines are clonal propagated to retain genetics - they accumulate mutations over time
high abundance of cell-layer specific mutations
What is heterozygosity? And why is it a challenge when it comes to grapevine genomics?
Heterozygosity is genetic variability between copies of the genome, i.e., many different alleles at numerous locations in the genome.
This is common in grapevines due to their long history of outcrossing and vegetative propagation
Challenging because:
Genome assembly complexity
high heterozygosity makes it difficult to assemble the genome accurately because the two haplotypes (maternal and paternal) can be very different. This can lead to fragmented genomes or incorrect assembly
Variant calling issues
distinguishing true variations from sequencing errors is more difficult when there’s a lot of natural variation in the genome
mapping and alignment errors
reads from one allele might not map well to a reference based on the other, reducing the accuracy of downstream analyses like gene expression
Can high heterozygosity be beneficial in grapevine genomics? If so, why or how?
It contributed to rich diversity seen in grapevine varieties - which is valuable for breeding and adaptation
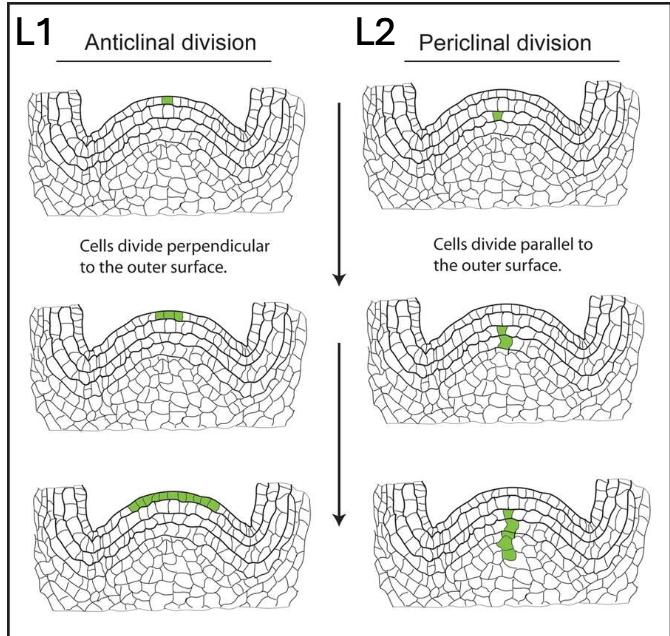
What is the structure of the grapevine meristem?
3 layers, but the two main ones are:
L1 (outer later) - develops the epidermis (skin of berry)
Anticlinal division, ie., cells divide perpendicular to the outer surface
L2 (sub epidermal layer) - contributes to internal tissues, including flesh and gametes (passed on through seed)
Periclinal division, i.e., cells divide parallel to the outer surface
Leaves and berries are L1 + L2 derived, whereas roots and woody shoot pith are only L2 derived
What is involved in L1 mutations?
affect only the outer epidermal layer
can result in changes to berry skin traits (colour, texture, bloom)
since L1 does not produce gametes, these mutations are NOT INHERITED through seeds BUT CAN BE PROPAGATED VEGETATIVELY (through cuttings)
Example:
Colour mutation in pinot varieties = classic example of L1 chimeras
What is involved in L2 mutations?
affect the internal tissues and reproductive cells
can alter traits like pulp composition, seed development and are heritable
more likely to be detected through whole-genome sequencing due to their impact on more extensive tissue types
Example:
heedlessness in some table grapes like Thompson seedless
caused by L2 mutations affecting development of ovules and seeds
What are the genes that control berry skin colour?
MYBA1 and MYBA2 which are found in chromosome number 2
They are the transcription factors for the last step in anthocyanin synthesis, where in the second copy of the gene there is an insertion of a retrotransposon “jumping genes” upstream of MYBA1 (green gene), resulting in a loss of function insertion or deletion in MYBA2 (blue gene with stars)
Only 1 functional copy of the MYBA locus is sufficient to produce anthocyanin, another example is chardonnay and Cabernet Sauvignon - where chardonnay has a retrotransposon and cab sauvignon does not - resulting in a coloured berry
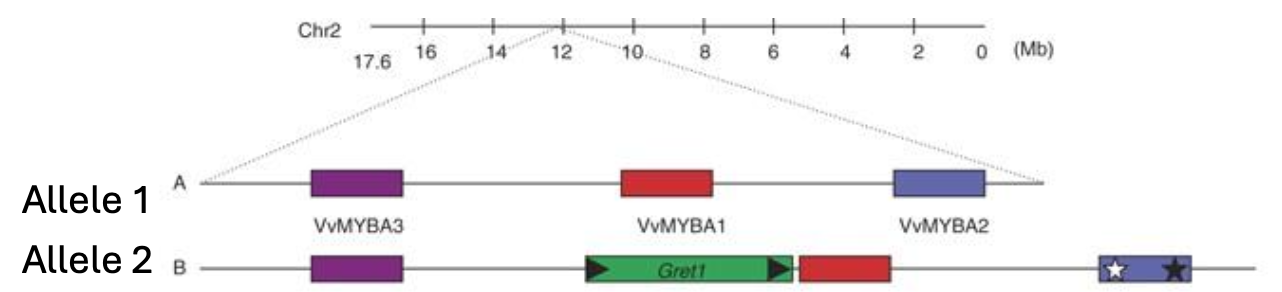

What makes the “gris” mutation?
Gris phenotypes only have one gene working in the L1 layer, MYBA1 or MYBA2 are either deleted or transcriptionally silenced
since the L2 layer remains similar to Pinot Noir, the internal flesh may still contain some anthocyanin, but this is masked by the pale skin - resulting in a pink colour
Can occur in different cultivars, most commonly known is pinot gris
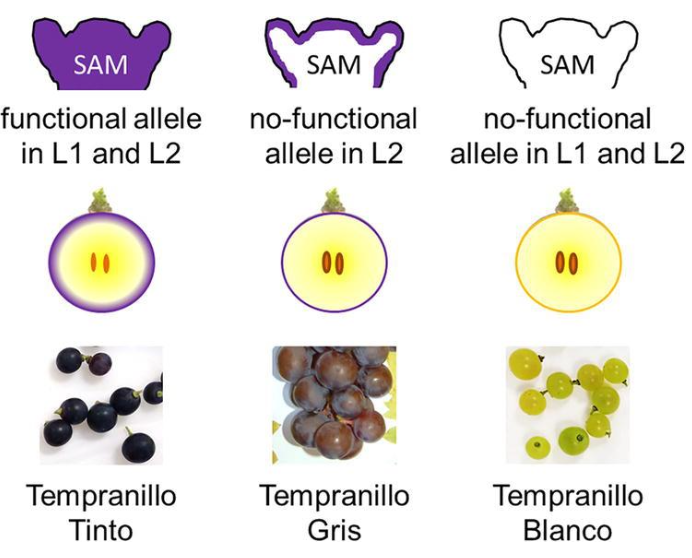
How are somatic mutations passed on?
Are not inherited through seeds (unless L2 is also mutated) but are passed on via vegetative propagation, which is why these colour variations are preserved as distinct cultivars

What makes the “blanc”mutation, in a variety like pinot blanc in comparison to Pinot Noir?
In pinot blanc, further mutation leads to complete inactivation or deletion of MYBA1 and MYBA2 in L1, and possibly also reduced formation in L2, resulting in no anthocyanin production and a white berry
Where as Pinot Noir has both MYBA1 and MYBA2 fully functional so we get a dark skinned berry

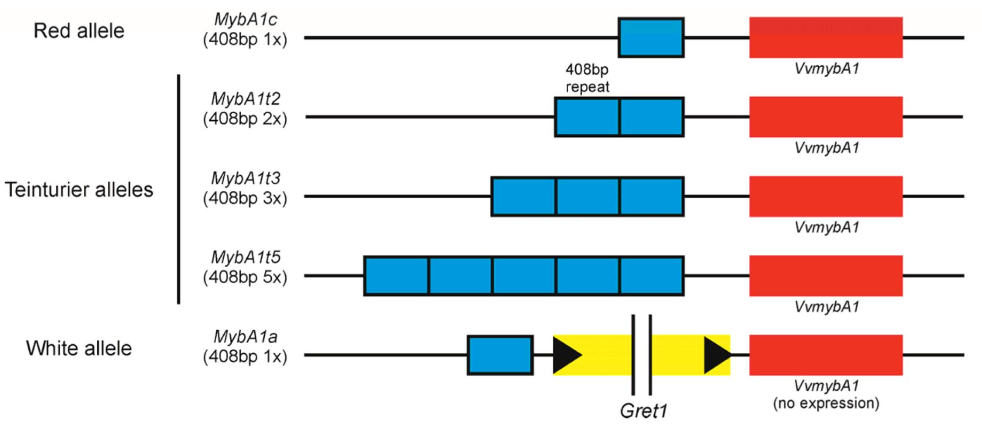
What is involved in “teintureir” berries? i.e., red flesh phenotype
The red colouration is due to the presence of anthocyanins in both the L1 and L2 layers.
L2 layer DNA is repeated which increases the expression of MYBA1
More copies of the DNA repeat = more anthocyanin
Image shows where the blue boxes are DNA copies
Where do muscat varieties (muscat of Alexandria, muscat gordo blanco) get their “muscat character” (floral, fruity)?
Due to high accumulation of monoterpenes - class of volatile compounds with low odour thresholds and strong floral notes
DXS is a key enzyme in the MEP pathway - that acts as a precursor for the synthesis of monoterpenes and diterpenes
PATHWAY SUMMARY:
DXS catalyses
DXP is then converted to IPP and DMAPP which are building blocks for monoterpenes and diterpenes
In muscat varieties, a specific point mutation in the vVDXS1 gene is strongly associated with increased monoterpene accumulation
This mutation increases the enzymes activity = greater terpene production
How is rotundone (pepper) aroma produced in shiraz?
2 different alleles of VvTPS24 producing different concentrations of a-guaiene (precursor for rotundone)
VvTPS24 = 3.5% a-guaiene
VvGuaS (two mutations compared to VvTPS24) = 44% a-guaiene
Shiraz only has one functional copy and is the high a-guaiene producing version
What wild grapevine species have vitis vinifera originated from?
Vitis sylvestris
How has genomics helped us trace the domestication of grapes?
in 2023 scientists sequenced 1604 V. vinifera and 844 V. sylvestris accessions (a distinct, uniquely identifiable sample of plants representing a cultivar) and produced a complete reference genome for V. sylvestris using long-reads
Genomics indicates the presence of 4 different V. sylvestris groups
The genetic diversity of all the sequenced V. sylvestris accessions shows the presence of 4 ecotypes - 2 in Western Europe and 2 in the east (caucasus and west Asia)
The eastern and western ecotypes separated ~200,000-400,000 years ago
Mapping of genetic similarity between V. vinifera and V. sylvestris groups reveals two domestication hotspots (CG1 and CG2), one in western Asia and Caucasus, dated 11,000 years ago. So wine grapes DID NOT predate table grapes
CG1 table grape early domesticates dispersed into Europe through Anatolia. Crossings with local V. sylvestris during dispersal helped CG1 table grapes adapt and become the known wine grape varieties we have today
What are clones?
Grapevine clones are genetically identical plants that comes from a single parent vine and are propagated vegetatively (for its desirable traits).
Small mutations can occur over time that lead to subtle differences in traits like: berry size, yield, flavour/aroma compounds, disease resistance, phenology
Because of this different clones of the same varieties can be chosen to suit specific climates, wine styles or vineyard goals
What are the problems with clones?
identity currently relies on record keeping - lots of room for human error
only 1-200 mutations separate clones in 500 million DNA bases
Why is it important to reliably identify grapevine clones?
bushfire recovery - obtain reliable clones for replanting material
insights into heritage material - relationship to modern clones, patterns of historical distribution
quality control of propagation material - mother vines and pre/post importation material