Mod 15 - Biotechnology: Recombinant DNA Technology and Forensic DNA Profiling
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
25 Terms
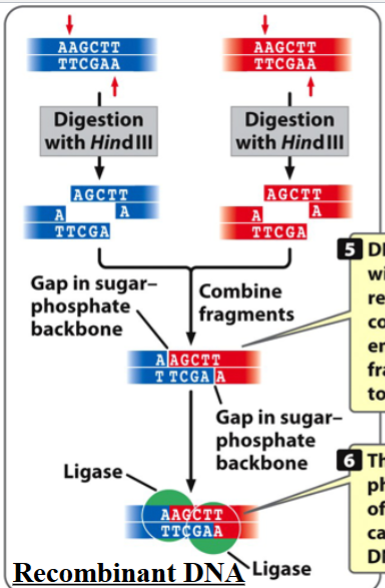
creating recombinant DNA molecules
putting together restriction fragments of DNA from different origins
DNA molecules cut with the same restriction enzyme have complementary sticky ends
those DNA molecules an pair if the fragments are mixed together
nicks in the sugar-phosphate backbone of the two fragments can be sealed by DNA ligase
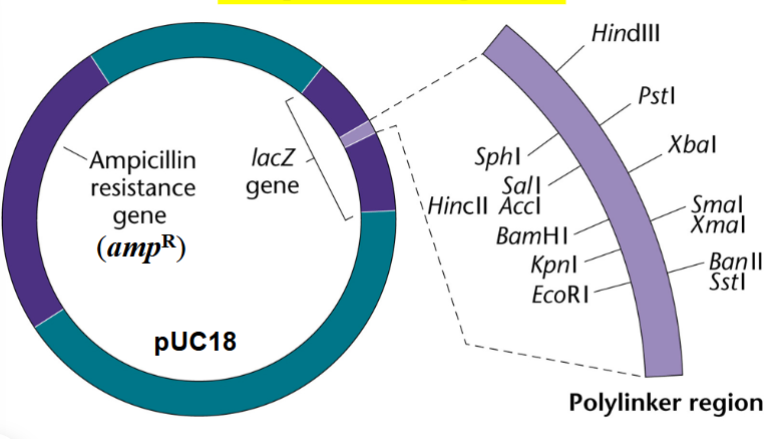
cloning vector
used to make copies of an isolated gene or of any fragment of DNA of interest
e.g. pUC18
components of the pU18 cloning vector
ampR: ampicillin resistance gene
lacZ+ gene: codes for beta-galactosidase (taken from lac operon)
^beta-galactosidase breaks down lactose and breaks down X-gal
polylinker region inside the lacZ+ gene is a cluster of restriction sites for a number of different restriction sites for a number of different restriction endonucleases
^any of the endonucleases may be chosen to insert a foreign fragment of DNA (e.g. an isolated gene of interest)
inserting any foreign fragment of DNA in the polylinker region would disrupt and disable the lacZ+ gene → make the plasmid lacZ-
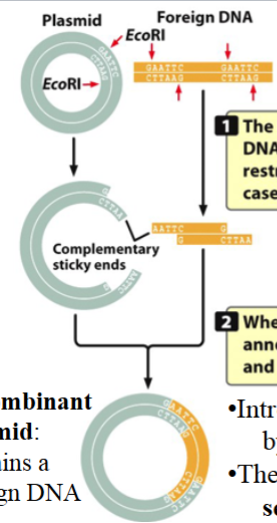
creating recombinant DNA molecules with a cloning vector
e.g. using pUC18 as a cloning vector:
plasmid is incubated with EcoR1 (a restriction enzyme)
plasmid is cut at the polylinker and linearized
foreign fragment of DNA containing a gene of interest is also treated with EcoR1 → will have the same sticky ends as the linearized plasmid
plasmid and foreign DNA mix → result is a recombinant plasmid
recombinant plasmid disrupts and disables the plasmid’s LacZ+ gene
stable recombinant plasmid is introduced into bacteria by transfection
as bacteria multiply, they also multiply the plasmids with them → many copies are made (cloned)
transfection
the process of introducing nucleic acids into cells by nonviral methods
used to introduce a stable recombinant plasmid into bacteria
antibiotic selection
a growth medium contains an antibiotic
only the bacteria that carry the correct antibiotic resistance gene will be able to survive
e.g. ampR (ampicillin resistance gene) in a plasmid, e.g. pUC18

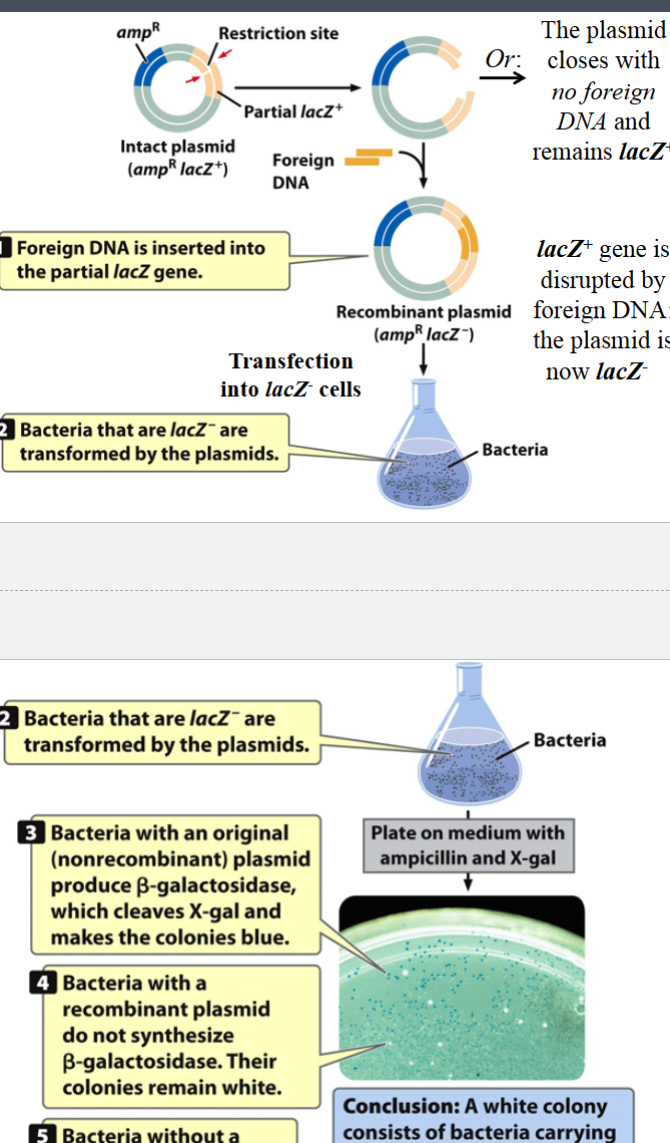
X-gal assay: antibiotic/blue-white double selection assay (an example of creating recombinant DNA molecules with a cloning plasmid)
used to identify E. coli colonies that contain cloned DNA fragments
beta-galactosidase can cleave X-gal (a lactose analog) into galactose plus an intermediate
the intermediate can be oxidized to turn it into an insoluble blue dye
host bacterium is a special strain of E. coli that is engineered to be lacZ- (does not have a lacZ+ gene → cannot make its own beta-galactosidase unless it picks up an intact pUC18 plasmid)
if the lacZ+ gene in the plasmid is intact, those cells will turn blue in an X-gal selection assay
if the lacZ+ gene in the plasmid has been disrupted by the insertion of foreign DNA in the polylinker region, those cells will not turn blue because they will stay lacZ-
bacteria with original (nonrecombinant) plasmid produce beta-galactosidase → colonies turn blue (X-gal positive)
bacteria with recombinant plasmid do not produce beta-galactosidase → colonies remain white (X-gal negative)
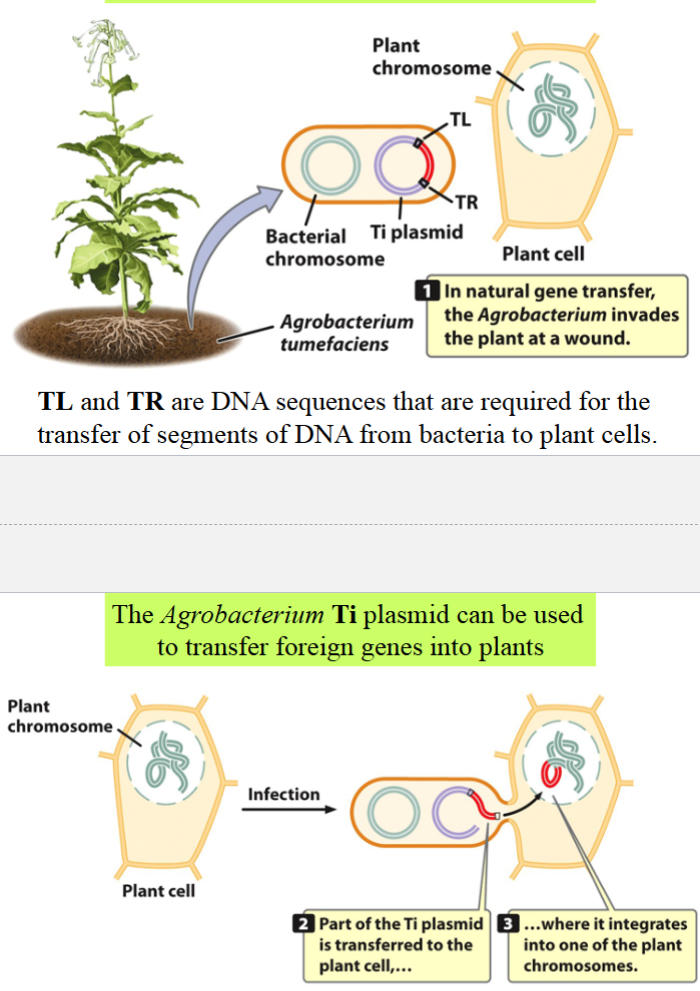
Agrobacterium Ti plasmid
can be used to transfer foreign genes into plants
in natural gene transfer, Agrobacterium invades the plant at a wound
TL and TR are DNA sequences required for the transfer of DNA segments from bacteria to plant cells
the segment of the Ti (tumor-inducing) plasmid that is flanked by TL and TR is transferred to a plant cell, integrated into its DNA
generating transgenic crops with glyphosate resistance
glyphosate: an herbicide that kills plants by inhibiting EPSP synthase
there is a fusion gene: EPSP gene is carried under control of a promoter from a plant virus in a Ti plasmid vector → EPSP synthase can be expressed in larger-than-normal levels in these plants
plant crops with this integrated fusion gene are resistant to concentrations of glyphosate that are normally fatal
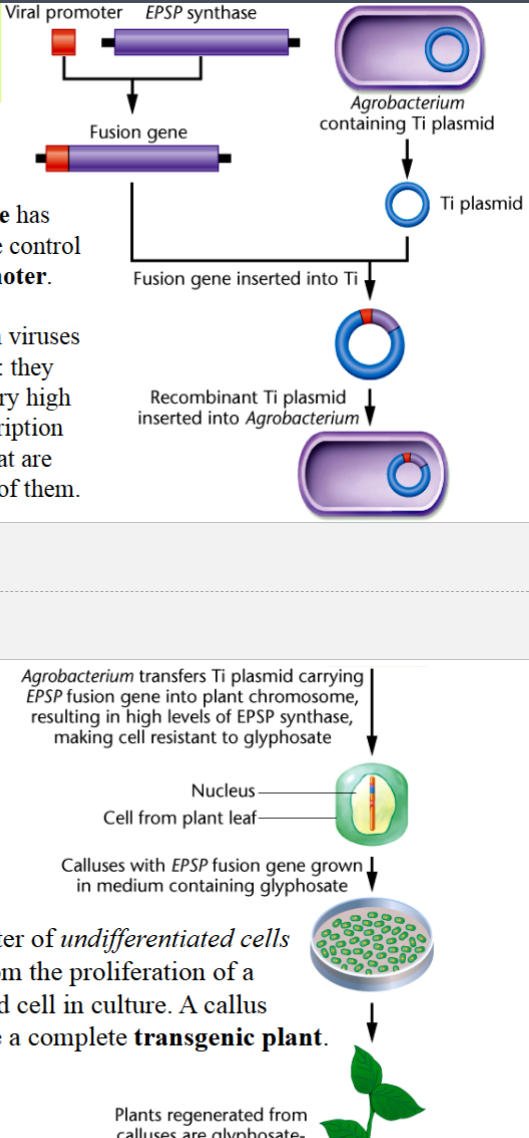
callus
a cluster of undifferentiated cells that results from the proliferation of a Ti-transformed cell in culture
can regenerate a complete transgenic plant
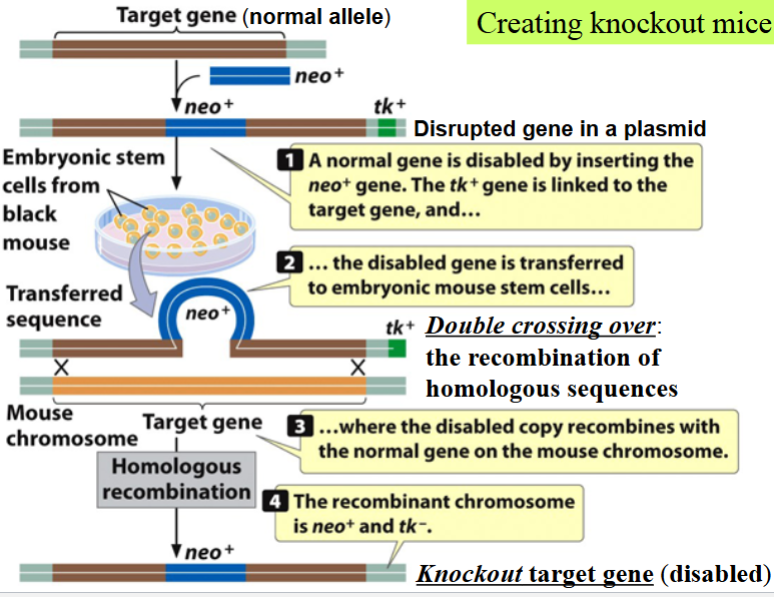
(transgenic) knockout mice: an example of gene targeting
a type of transgenic mice
generated after the homologous recombination of a cloned mutant allele with the target cellular gene of interest in embryonic stem (ES) cells

generation of knockout ES cells
cloned gene of interest (target) is disrupted with a segment of DNA containing neo+ (neomycin resistance gene) → makes the target gene a null mutant allele
disrupted gene is in a plasmid vector also containing tk+ (thymidine kinase gene) from the herpes simplex virus
(thymidine kinase can phosphorylate the thymidine analog gancyclovir → turn it into a toxic inhibitor of DNA replication → kill cell)
plasmid is introduced into mouse ES cells
things that may happen in knockout ES cells
knockout or random integration
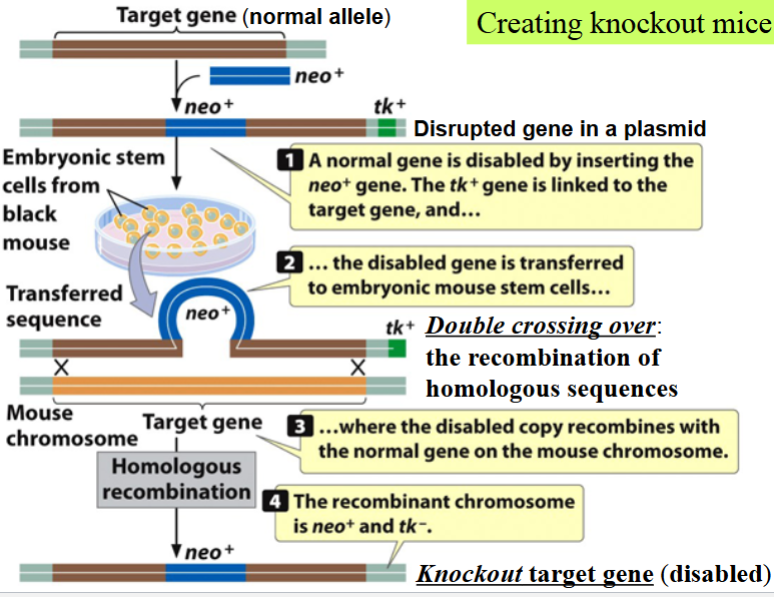
knockout in mouse ES cells
homologous recombination (by double crossing over) of disrupted target gene in the plasmid with the cellular version
a copy of the normal cellular target is replaced with the disrupted version
these cells will be able to grow in both neomycin and gancyclovir because the tk+ gene will not be integrated into their genomes
random integration of the entire plasmid vector in mouse ES cells
happens by single crossing over into the chromosome of the cellular genome without knocking out the normal cellular target
these cells will not be able to grow in medium containing gancyclovir because tk+ gene will be integrated and expressed → toxicity is resulted
creating knockout mice
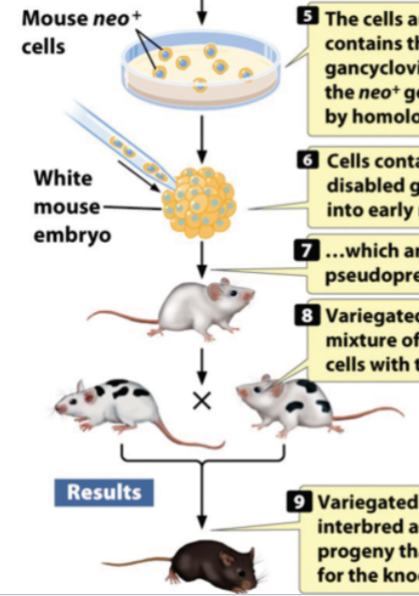
once created, knockout ES cells are injected into mouse embryos
knockout ES cells (neo+/tk-) from a black mouse are injected into early embryo of a white mouse
hybrid embryo is implanted in a foster mother
embryo develops into a chimeric mouse; may have black mouse germ cells
chimera mice are mated; some of the progeny will be homozygous knockout mice (black)
forensic DNA profiling
the use of genetic markets to distinguish individuals and answer questions to the legal system
the genetic markets are non-coding polymorphic loci
e.g. RFLPs, STR loci (aka PCR loci)
electrophoretic profiles from both methods above yield DNA fingerprints unique to every individual
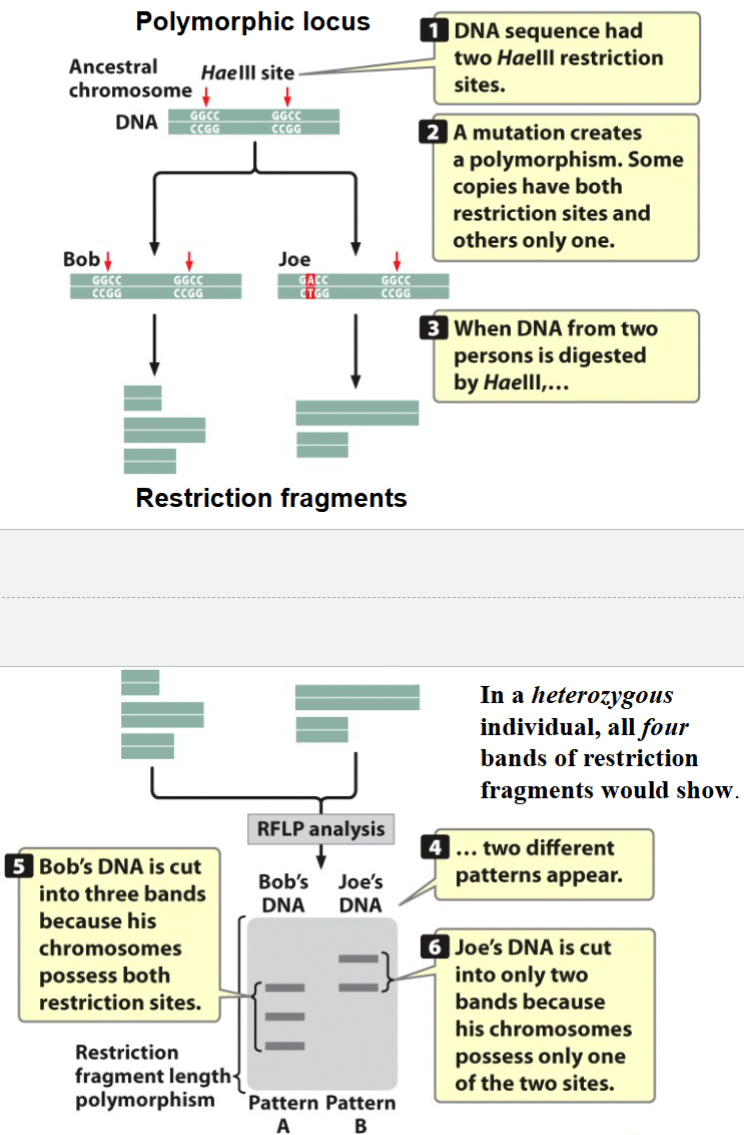
RFLPs
restriction fragment length polymorphisms
loci that vary in their numbers and positions of restriction sites
method is now outdated
an example of DNA evidence
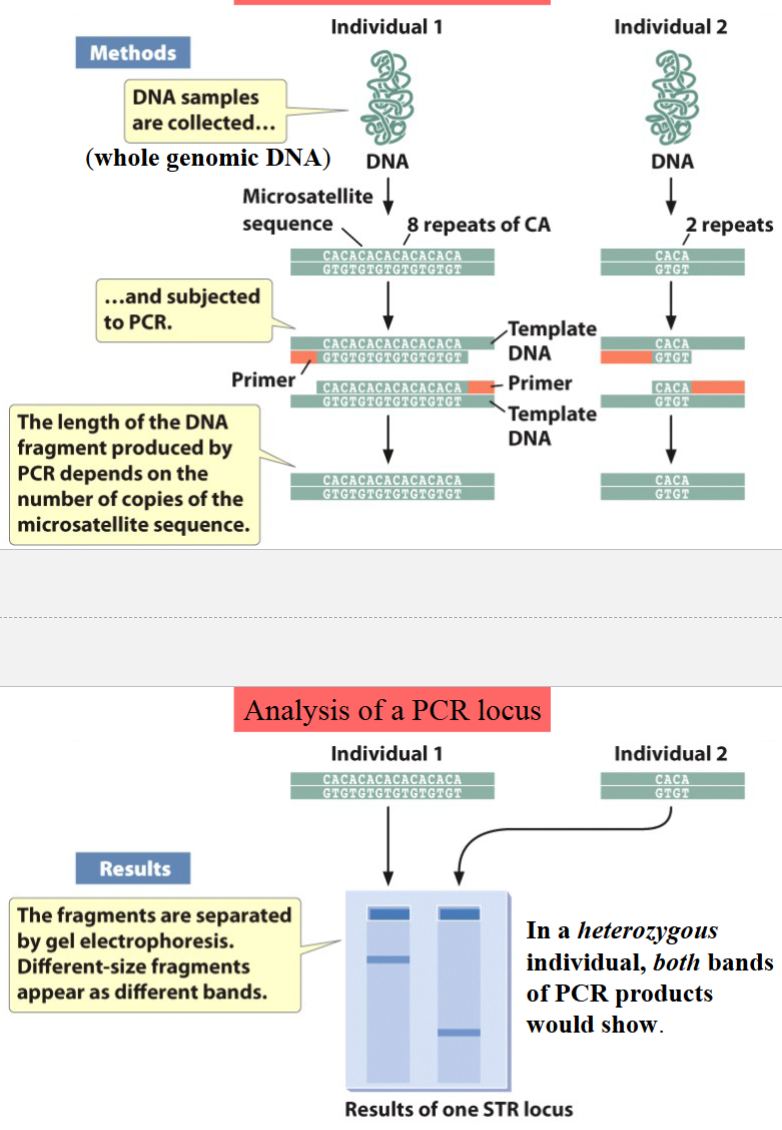
STR loci (aka PCR loci)
short tandem repeats
loci contain variable numbers of repeats of short sequences (microsatellites) in tandem
microsatellites (aka simple repeats) are 2 to 5 nucleotide sequences
the number of repeats in each polymorphic locus varies among individuals
analysis can be done using PCR, hence the name
an example of DNA evidence
e.g. D7S820, one of the 13 STR markers used by the FBI; number of GATA microsatellite repeats may be different in different individuals
example of STR locus/PCR locus
a single pair of primers targeting a single locus is used
both individuals are homozygous for this locus
result of PCR and electrophoresis of the product is a single band for each individual (corresponding to a single PCR product)
the primers used anneal just outside the region of the locus containing the microsatellite repeats → the length of the PCR products depends on the number of repeats of the microsatellite sequence (CA, in this case)
if an individual were heterozygous, both bands of PCR products would show
if multiple pairs of primers were added to the PCF mixture (targeting several PCR loci at once), the pattern of bands would be a unique DNA fingerprint (see slide 34 for an example)
CODIS
combined DNA index system
the core of the national DNA database
established by the FBI
developed to enable public forensic DNA laboratories to create searchable DNA databases of authorized DNA profiles
(not important for the test, just cool extra info)
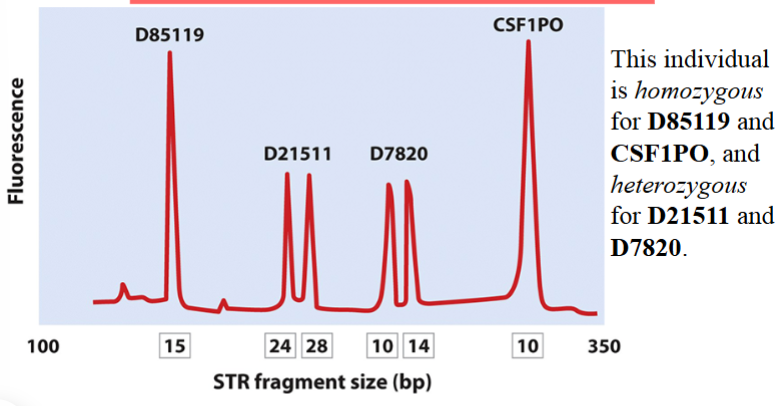
DNA fingerprinting with multiple PCR loci
primers used for fingerprinting can be tagged with fluorescent labels → resulting PCR products can be detected with a laser beam
as products are separated by electrophoresis, the laser beam detects them as they migrate past it → represented as peaks
homozygous loci will appear as a single peak
heterozygous loci will appear as double peaks
the amelogenin test
amelogenin: a protein found in developing tooth enamel
amelogenin is not an STR locus, but can be analyzed with PCR
gene (AMEL) is present in both X and Y chromosomes, but the X-linked version is shorter
can be used to determine the sex of human DNA samples through PCR of intron 1 of the gene
PCR of AMEL-X gives rise to a 106-bp product (shorter)
PCR of AMEL-Y gives rise to a 112-bp product (longer)
examples of DNA evidence in forensic identification
identification of 9/11 victims: using PCR from small samples (fingertips, e.g., matched with DNA from razor blades, combs, toothbrushes)
OJ Simpson trial: DNA from suspect found on scene, DNA from victim found on suspect’s shoe
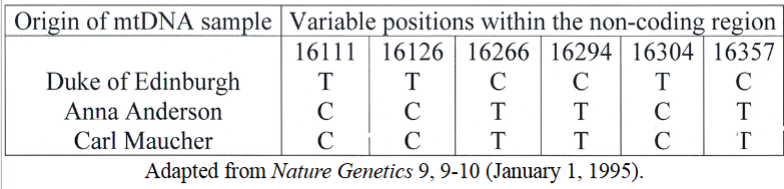
Romanov family and Anna Anderson: mitochondrial DNA
mitochondrial DNA forensics
used in the case of Anastasia Romanov and Anna Anderson
Anderson claimed she was Anastasia
mitochondrial DNA (mtDNA) from Prince Philip’s hair was taken (because Philip was related to Anastasia maternally)
Anderson’s mtDNA did not match Philip; matched a relative of Franziska Schankowska
polymorphic sites in mtDNA are single-nucleotide positions (picture demonstrates how Anderson matched Schankowska but not Philip and thus did not match Anastasia)