microbio lecture 24 (microbiomes)
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
microbiota
the microorganisms inhabiting a specific environment/habitat
microbiome
the collective gene pool of the microbiota (are microbial ecosystems)
carl woese
discovered 16S used to build a molecular clock AND identify unculturable microbes (how we study microbiomes)
study microbiome methods
16S rRNA amplicon sequencing
metagenomic sequencing
transcriptomics/RNAseq, metabolomics and proteomics
16S rRNA amplicon sequencing
way we study microbiomes:
identifies which microbes are there
metagenomic sequencing
way we study microbiomes:
identifies which microbes are there and what functions they have
transcriptomics/RNAseq, metabolomics and proteomics
way we study microbiomes:
identify what functions microbes are actively doing
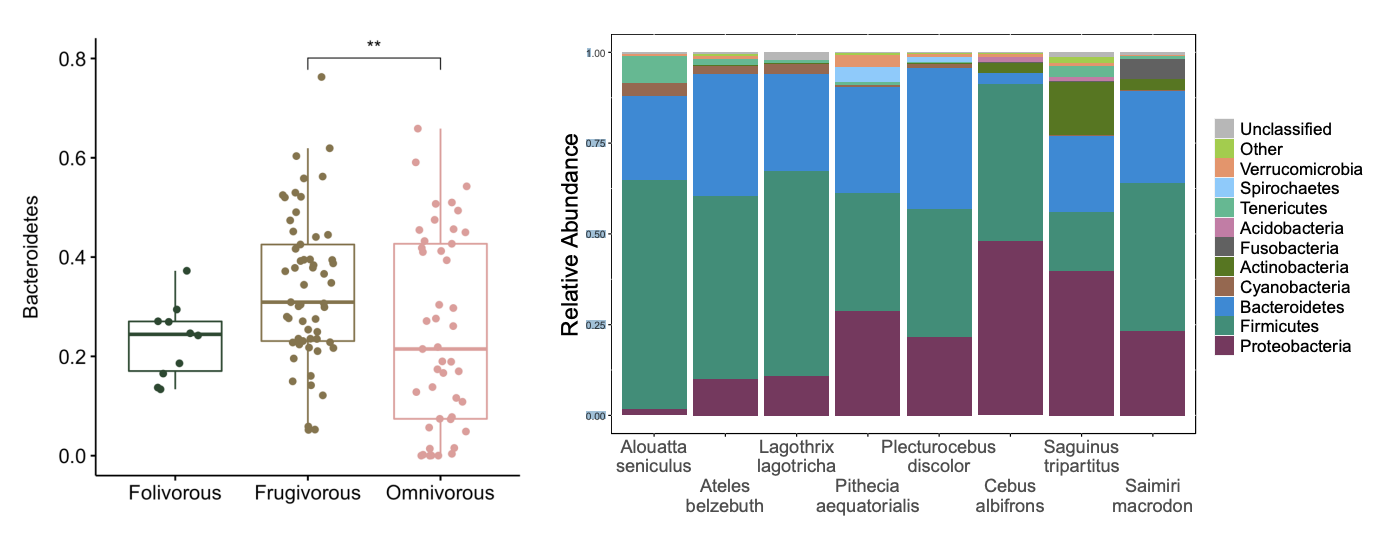
alpha diversity, beta diversity, taxonomic abundance
We examine differences in microbial communities using three common metrics:
alpha diversity
Within-sample diversity
How diverse are the microbial taxa the microbial taxa in a single sample?
Species richness – the number of microbial taxa
Species diversity – the evenness of microbial taxa
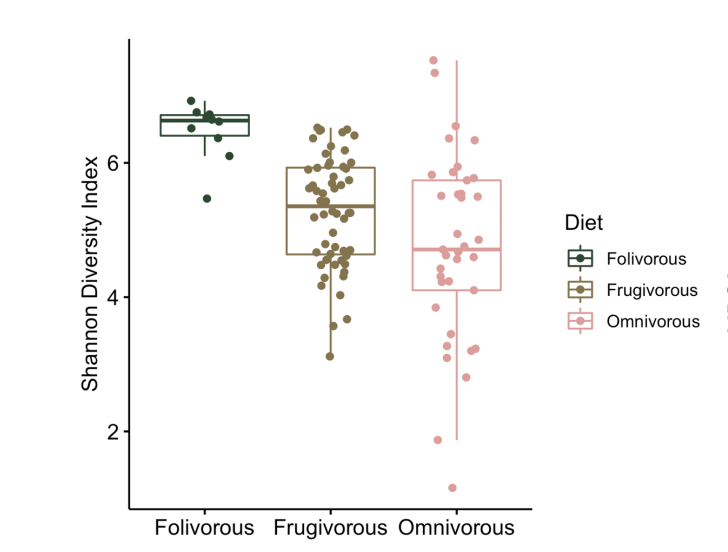
beta diversity
Between-sample diversity
How do microbial communities differ between samples?
Community level similarity or dissimilarity
How much overlap in species composition is there between two samples?
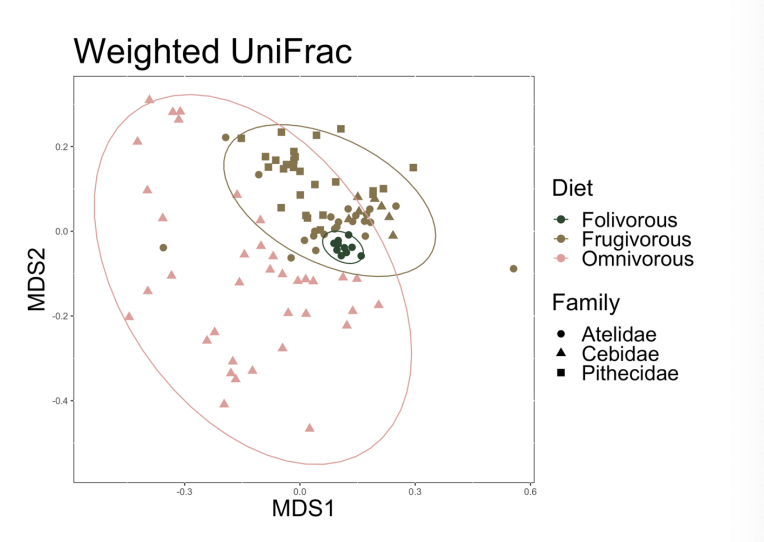
relative abundance
what proportion of our sequences are assigned to a taxa
where do we study microbiomes
Virtually every habitat on earth:
Water (marine and freshwater)
Air
Soil
Plant surfaces
Animals
Thermal vents
Buildings
human microbiome project goals
Determine whether individuals share a core human microbiome
Understand whether changes in the human microbiome are correlated with changes in human health
Develop the new technological and bioinformatic tools needed to support these goals
Address the ethical, legal and social implications raised by human microbiome research
human microbiome project sample collection
Microbiome samples collected from 300 healthy individuals
Initially, to characterize the complexity of the microbial communities, samples were subjected to 16S RNA sequencing
Metagenomic sequencing was used to provide insights into functions performed by the human microbiome
human microbiome
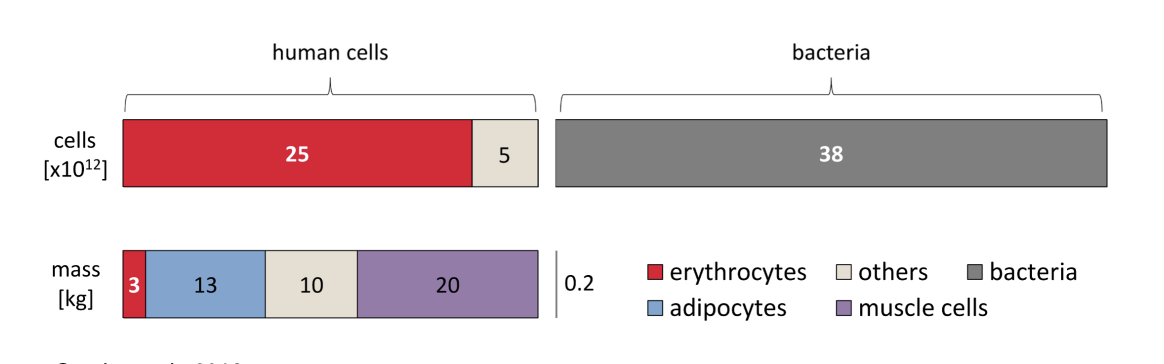
Humans have more microbial cells than human cells (~1.3:1 ratio)
Extensive variation within and between people
Alterations in the microbiome are associated with health and phenotype
Microbiome-based therapies are being developed
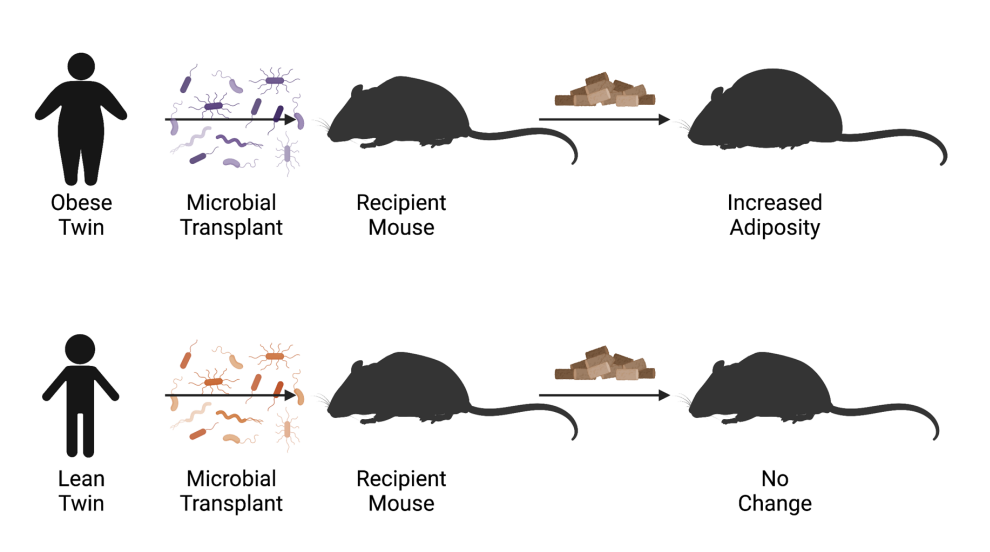
phenotype
our microbiome influences our ______:
fecal transplant from lean twin conferred a lean ______
fecal transplant from obese twin conferred an obese _____
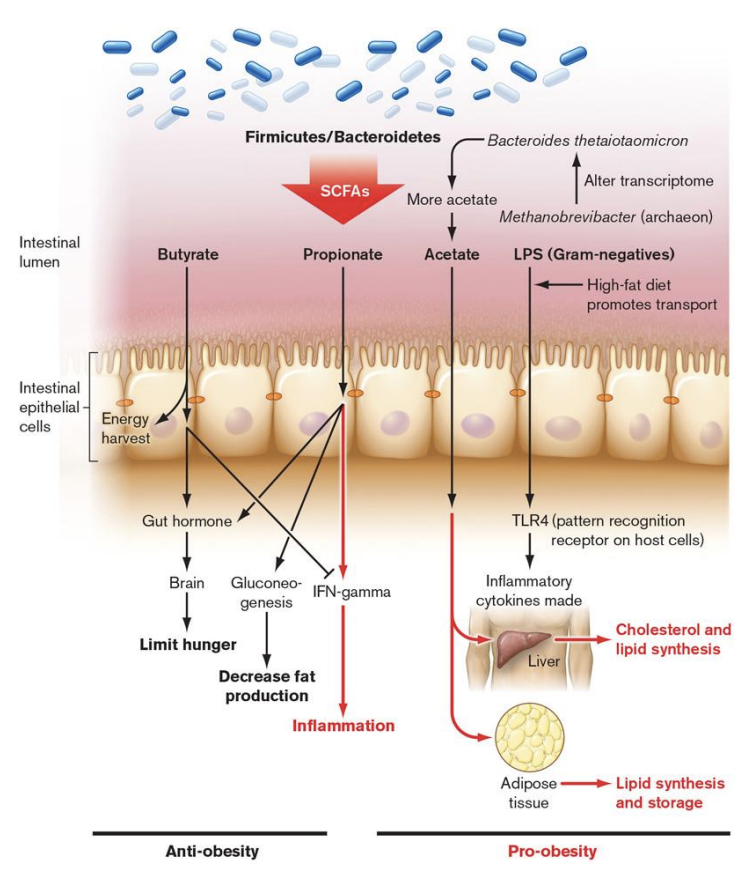
short chain fatty acids (SCFA)
Our microbiome influences our phenotype – How?
through ________: by-products of microbial fermentation
Some trigger anti-inflammatory immune pathways (butyrate and sometimes propionate)
gut microbiome
this develops rapidly during first 3 years of life
geography, lifestyle and parasite infection status ALL were associated with ___ _____ composition
we have evolved with our microbes
diet, environment and disease exposure all shape both our genome and our microbiome
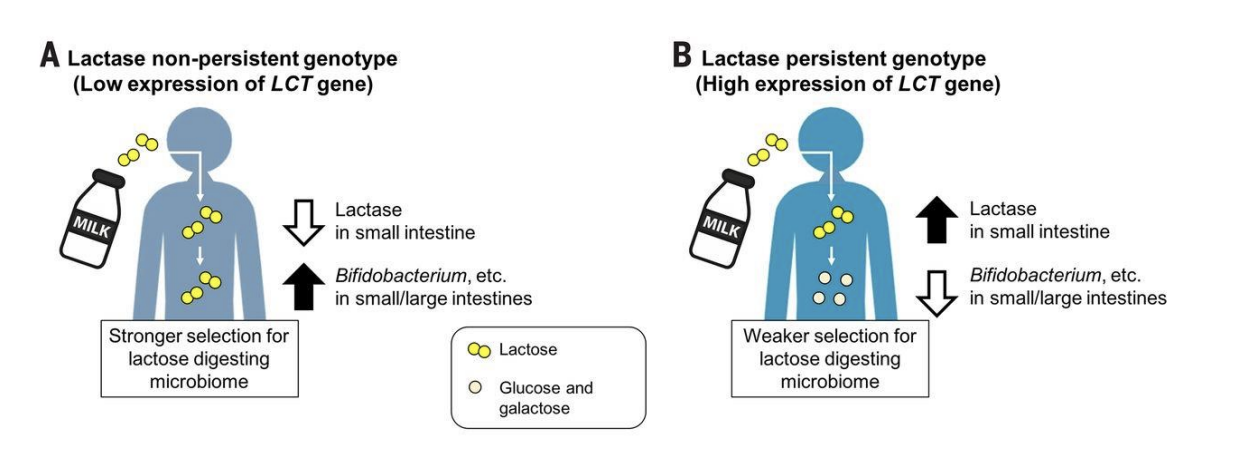
ex: low lactase (low expression of LCT gene) = higher bifobacterium in intestines
high lactase (low expression of LCT gene) = lower bifobacterium in intestines
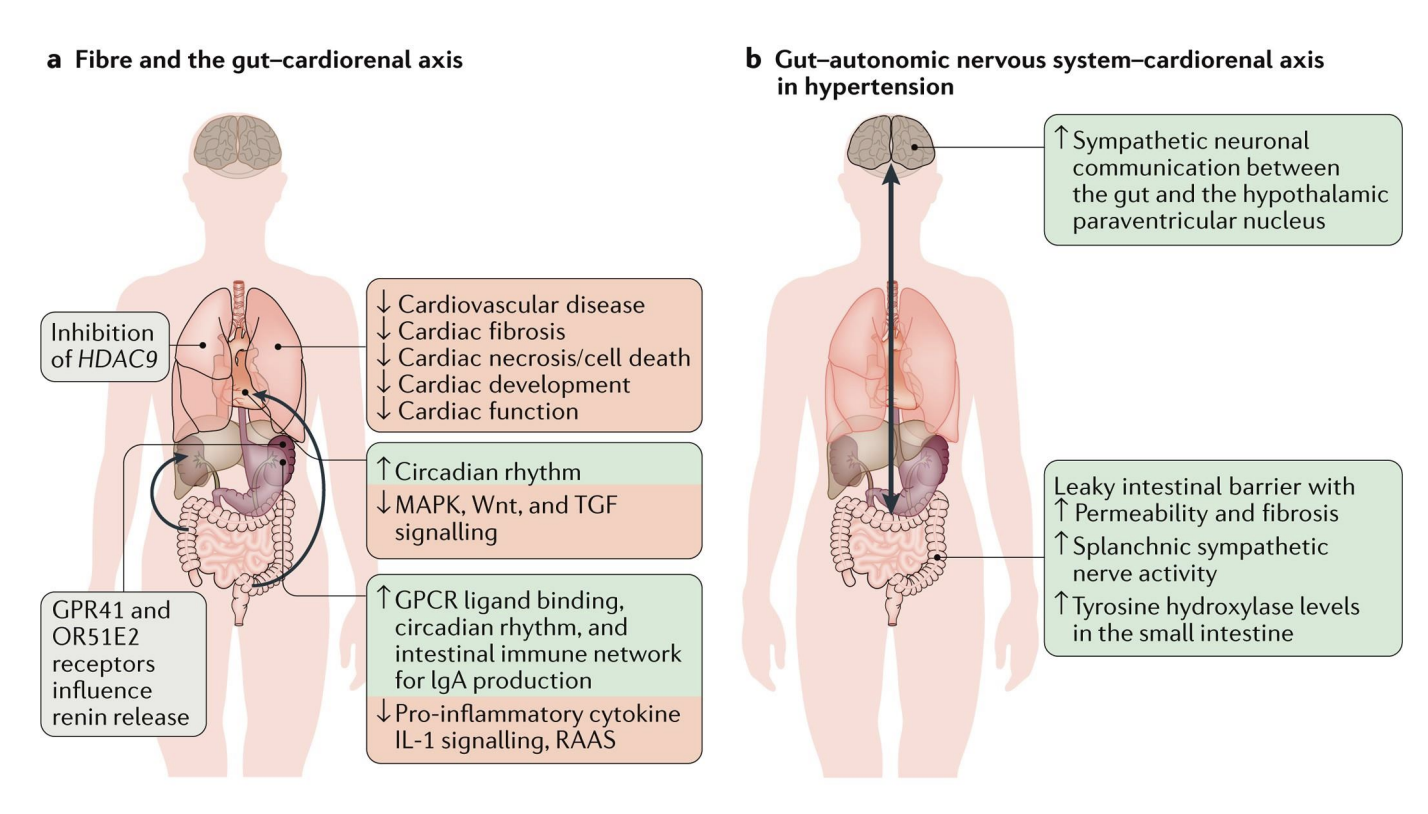
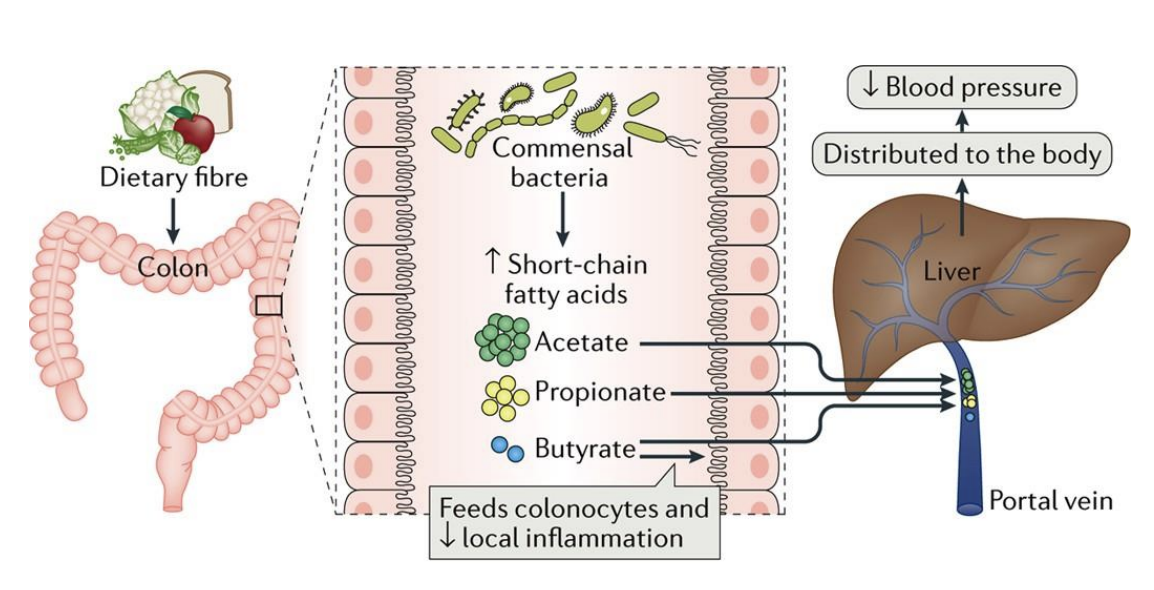
gut microbiome and hypertension
There is solid evidence that the gut microbiome plays a direct role in modulating blood pressure
association has roots in infancy
probiotic
foods and supplements containing live microorganisms
prebiotics
food containing substrates for beneficial microbes (usually fiber)
fecal microbiota transplant
transferring the microbiota from a health individual to a diseased individual
Highly effective for C. difficile infections
Not reliably effective for other diseases (and can be harmful)
toxins
Just as we use microbes to bioremediate toxic waste, many species rely on their gut microbiome to neutralize ___ in their diet
Giant pandas have gut microbes that degrade cyanide, a toxin found in bamboo
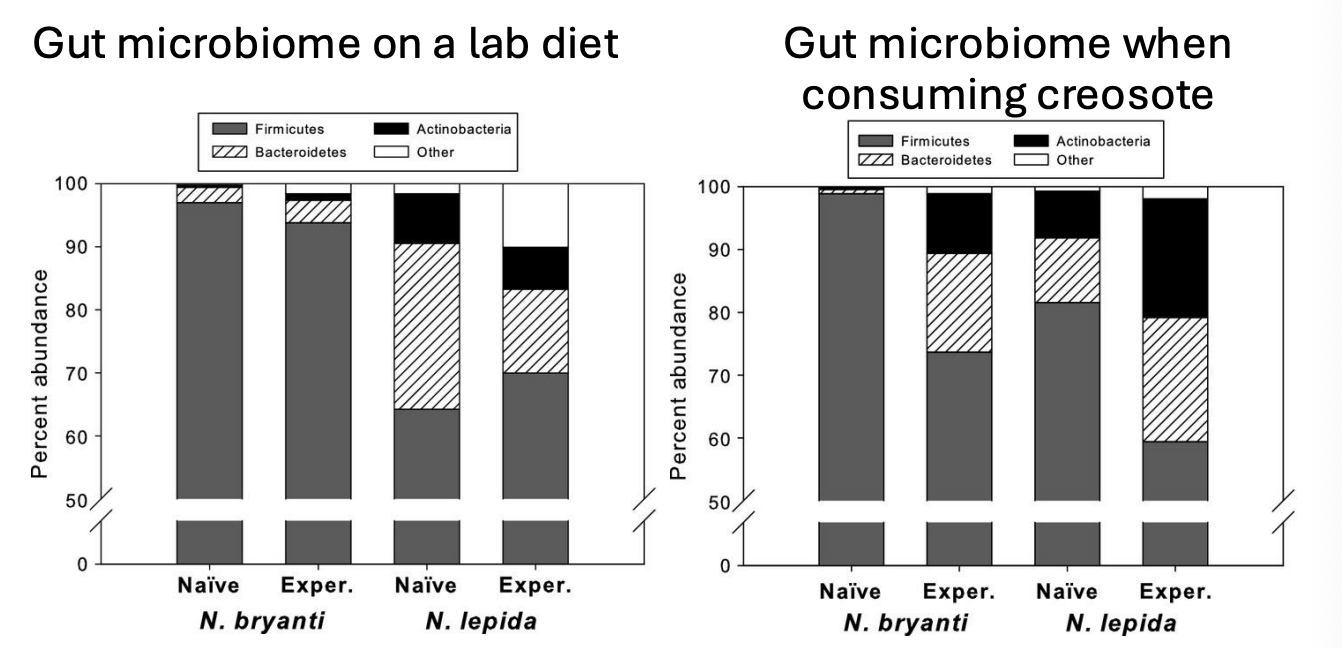
Pack rats (Neotoma spp.) have gut microbes that degrade creosote toxins
neotoma
Two different species of pack rat consume creosote bushes (Neotoma bryanti and N. lepida) in a portion of their range
rhizobium
is only one bacteria with a key role in plant-microbe interactions – there is an entire ecosystem of soil microbes
A robust soil microbiome:
Provisions plants with nutrients
Helps exclude pathogens
Soil microbiomes are affected by farming practices
built environment
microbiome of ____ ____:
Our human propensity to alter our environment has changed our interactions with microbes
This changes which microbes we acquire from the environment
May contribute to the decreased diversity of human-associated microbiomes in North America and Europe
→ The chemicals we use in building materials and in our daily lives affect the microorganisms around us
stress
Microbes in built environments experience more ___
UV light exposure
Low humidity
Temperature variation
Chemical exposure
Increased stress may induce the transfer of mobile genetic elements, facilitating the spread of antibiotic resistance
Additionally, increased stress selects for bacteria with resistance to stress (including resistance to antibiotics)
antibiotic resistance
Increased stress may induce the transfer of mobile genetic elements, facilitating the spread of _____ _____
Additionally, increased stress selects for bacteria with resistance to stress (including resistance to antibiotics)