CORSO- powerpoint 5 - EXAM 1
1/56
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
57 Terms
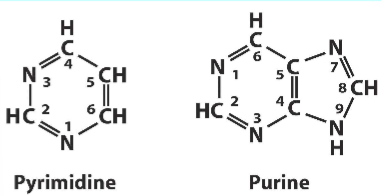
What is the difference between a pyrimidine and purine?
pyrimidine- 1 ring
purine- 2 rings
What’s the difference between names like adenine and adenosine? Cytosine and Cytidine? Guanine and Guanosine? Thymine and Thymidine? Uracil and Uridine
the difference is that adenine, cytosine, guanine, thymine, and uracil are THE BASE ONLY, when you ADD RIBOSE THEY turn into adenosine, cytidine, guanosine, thymidine, and uridine
What’s the difference between a nucleoside and a nucleotide?
A nucleoside doesn’t have a phosphate. (nucleotide= phosphate + nuceloside)
What’s the difference between DNA and RNA?
DNA uses deoxyribose, RNA uses Ribose—> there is a 1 oxygen difference
In addition to DNA and RNA, what else are nucleic acids used for?
cAMP
CoA
ATP
FAD
NAD+
DNA is how many strands? What bases pair with each other?
DNA is 2 strands
A pairs with T
C pairs with G
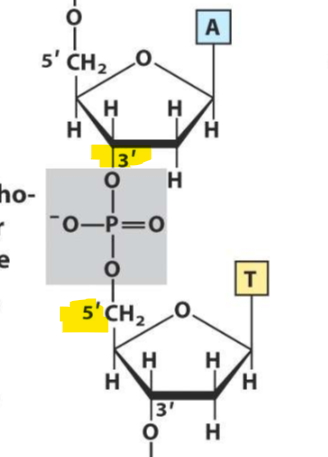
DNA and RNA is synthesized in the___’—>___’ direction
5’—> 3’
Where is the phosphate attached to on the bases of DNA and RNA?
Since DNA and RNA goes in the 5’—>3’ direction, The phosphate is attached to the 5’ OH group on the ribose before and the 3’ OH group on the ribose after it
What is the primary and secondary structure of DNA?
primary- amino acid sequence
secondary- double helix
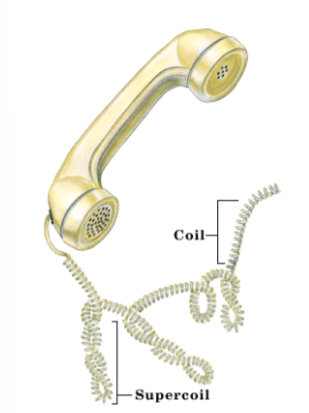
What is the tertiary and quaternary structure of DNA?
tertiary- the double helix that is SUPERCOILED
quaternary- DNA wrapped around histone proteins/ thickness of the chromosome
What are histone proteins? Why are they attracted to DNA?
proteins that are HIGHLY positively charged (bc of lysine and arginine), attracted to DNA because DNA is negatively charged
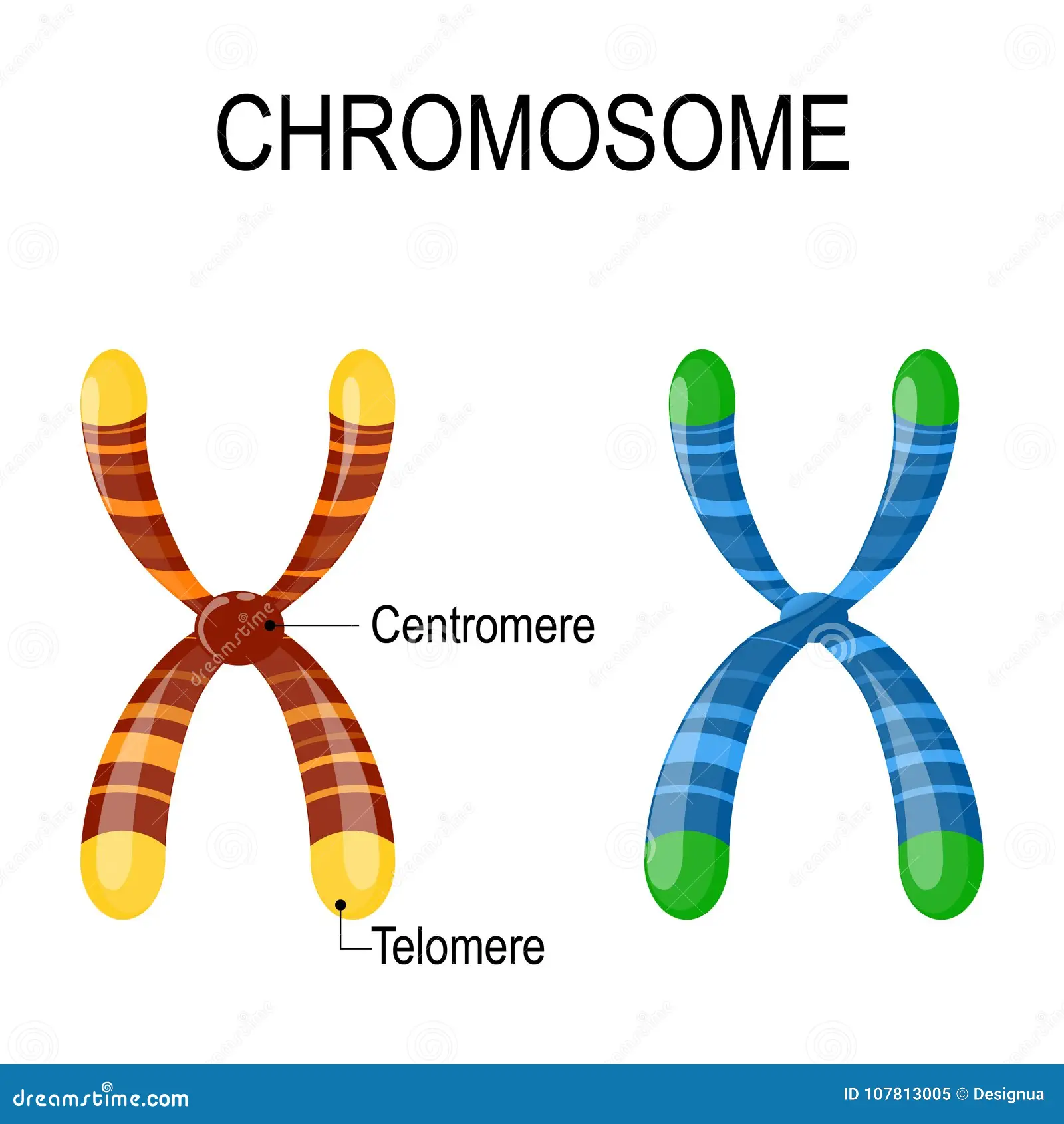
What are centromeres and telomeres?
centromeres- point of attachment between two chromosomes after DNA duplication
telomeres- protective caps on linear chromosomes (protect chromosome from degredation)
What are the stages of the cell cycle? When does the cell divide? When is DNA synthesized?
M- MITOSIS aka CELL DIVIDES
G1
S- DNA SYNTHESIS OCCURS
G2
repeat steps
How do G0 and GTD interact with the cell cycle?
G0- contact inhibition? basically a nondividing state
GTD- terminally differentiated aka no more cell cycle
What is the importance of the restriction point in the cell cycle?
to avoid mitotic catastophe
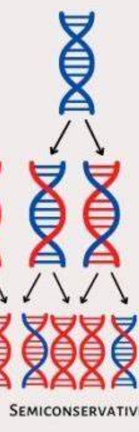
What does semiconservative replication mean?
when we replicate 1 strand of DNA is new and 1 is old
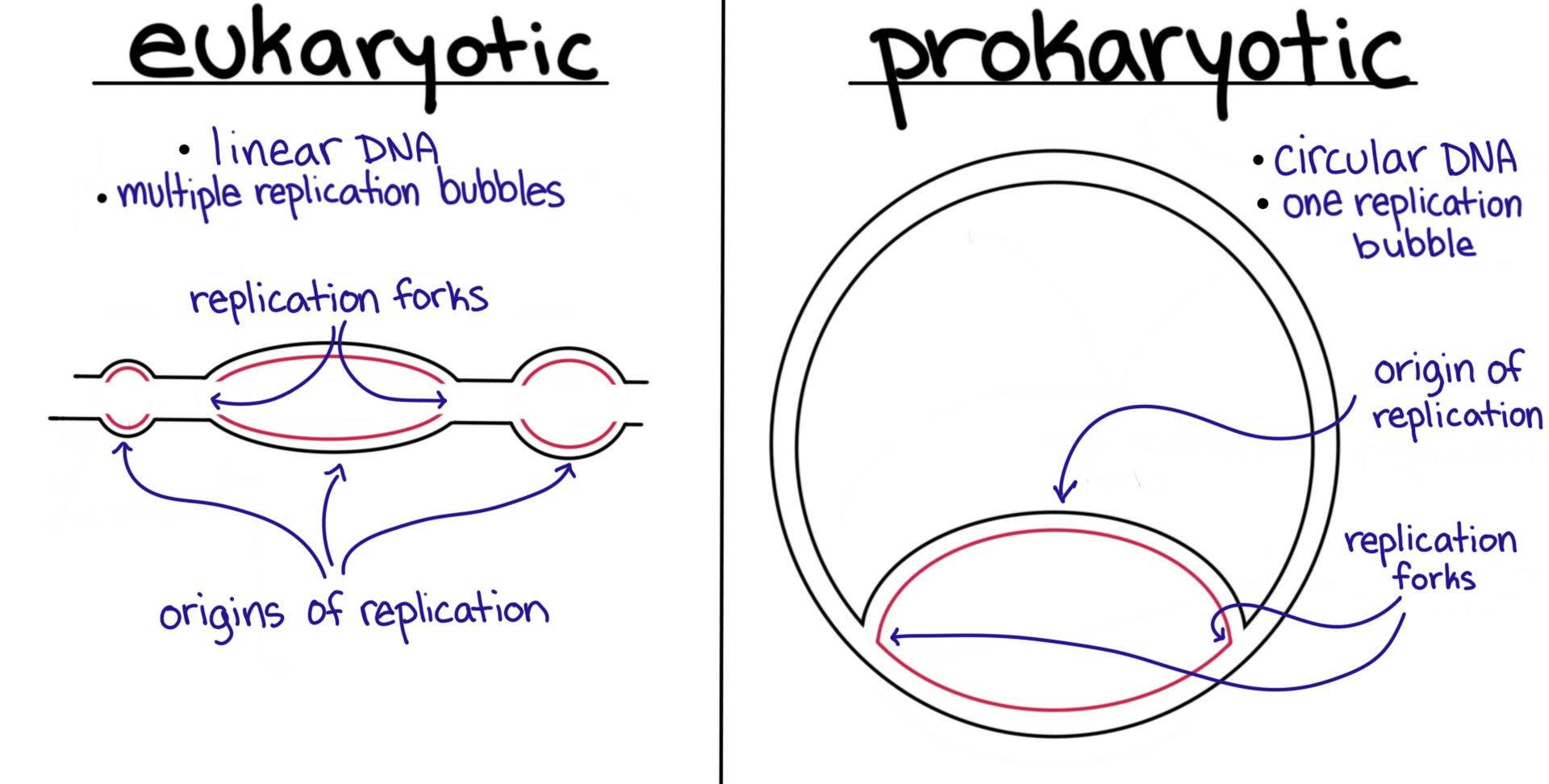
What is the OriC site? What is the OriC site high in?
aka the origin of DNA replication or where DNA replication starts—— high in A-T pairs to make it easier to unzip
How many replication forks does the OriC site have?
2
What is a replisome?
collection of enzymes at the replication fork
How does the OriC site on eukaryotic DNA compare to prokaryotic DNA?
in prokaryotes- one OriC site
in eukaryotes- multiple OriC sites
What are topoisomerases and what is the difference between type 1 and type 2?
they are needed to uncoil DNA before replication and then recoil after
difference between type 1 and type 2
type 1- break one strand and change linking number by 1
type II- break both strands and change linking number by 2
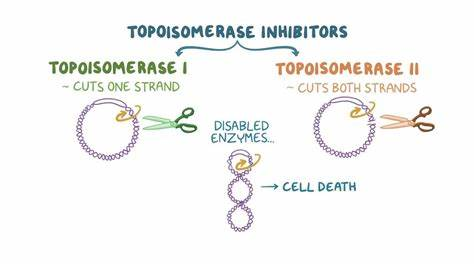
IN CANCER—→ WHY DO WE WANT TO INHIBIT TOPERISOMERASES?
said “possible exam question”
they can function to suppress rapidly dividing cells
What is a replication fork?
where we separate DNA in order to synthesize new DNA
Since DNA is ONLY synthesized in the 5’ to 3’ direction, what problem does this create?
creates a problem for the lagging strand that is anti parallel so it has to be synthesized in fragments
What is a primer? What is a primer made of?
MADE OF RNA
used to START DNA synthesis
basically these RNA primers about 10-60 are put on the unzipped DNA before DNA polymerase comes in and puts down new DNA bases
What is an Okazaki fragment?
basically how we synthesize the lagging strand
we make short fragments of DNA aka OKAZAKI FRAGMENTS and they are later joined together to make the new DNA strand <3
How are primers removed?
DNA polymerase I
How does the cell know which strand of DNA was the original strand?
once in awhile we add an extra methyl on DNA, when DNA replication occurs the new strand is not methylated
What is methyl directed mismatch repair? What is the only sequence methyl’s are found on? Is the new strand or old strand methylated?
(i’m very sorry my explanation is long I just want to explain it well)
BASICALLY the problem is that we have a mismatched base (ex: A—> G). This methyl mismatch repair process will FIX the “mismatched” bases. We assume that the old strand has the correct base (A), and the new strand has the wrong base (G, when it should be T). However, we need to be able to tell which is the old and which is the new strand in order to fix the problem and that’s where the methyl on the old strand comes in. The NEW strand does NOT have this. Therefore, once we can tell which strand is which we can go in, form a loop around the wrong base, nick it, and replace it with the right base!!!!!!!!!!!
only sequence—> GATC//// methyl is attached to the adenosine (A) of this
OLD STRAND IS METHYLATED
How does the cell know if DNA is paired correctly?
Only A=T and C=G have the correct distance between them. Other combos can hydrogen bond but the distance is different and recognized as incorrect
When does methyl mismatch repair take place during the cell cycle?
S phase or DNA synthesis
Mismatch repair defects are most associated WITH WHAT CANCER???
said “TEST QUESTION”
colorectal
All repairs in DNA must be made before the…
restriction point (right before S phase)
What is deamination and how does it cause mutations? What are some agents that promote deamination?
said “possible test question”
what is it? removal of nitrogen from a base
how does it cause mutations? turns it into the wrong base
agents- NITROSAMINES
What are alkylations?
add a extra methyl (CH3) group
What is depurinations?
basically we remove the purine group (which we know has 2 rings)
Depurinations, dealkylations, and deamination’s are all examples of _____________ mutations.
chemical
What type of repair is for right after DNA replication?
methyl mismatch repair
What type of repair is to correct chemical mutations (smaller mutation)?
base-excision repair
What type of repair is to correct larger mutations?
nucleotide-excision repair
What type of repair is used to reverse mutations?
direct repair
Explain how base excision repair works:
we have a chemical mutation to one of the bases (depurination, alkylation, etc.)
we remove the base
we remove the DNA backbone
DNA polymerase I comes in and adds new DNA to repair
seal the nick
The biggest mutation that deals with nucleotide excision repair is pyrimidines like thymine being mutated into _____________.
dimers
What disease can be caused by a defect in nucleotide excision repair and causes EXTREME sun sensitivity, leading to high skin cancer risk and other problems?
Xeroderma Pigmentosum
Direct repair is simpler than other types of repair but requires more ___________.
UV light
explain direct repair:
Purpose: Reverses specific types of DNA damage directly, without excision
examples: repairs thymine dimers using UV light and alkylation’s using ENZYMES
What is cross over repair? What is another name for crossover repair?
also known as “recombinational repair”
we have 2 chromosomes—> if one is damaged beyond repair, we can use the code from the good chromosome, cross it over, and duplicate it
What genes are associated with cross-over repair? If defective what cancers are associated with each?
BRCA 1
breast cancer, cervical
BRCA 2
What gene prevents the cell cycle from going into the S phase when elevated?
p53 gene (a tumor suppressor gene)
What are the role of cyclins?
proteins necessary to pass restriction points
What’s the difference between necrosis and apoptosis?
necrosis- “crush/break/damage”—> exposure to toxin or physical damage—> membrane damage, cell swelling and lysis
apoptosis- programmed cell death—> cell is not needed—> usually no lysis
What factors can trigger apoptosis?
TNF
growth factors
radiation/toxins
What is the Ames Test? What does it correlate?
said test question
TEST FOR MUTANOGENECITY
Correlation with cancer is 90%
What is an allele?
different variations of the same gene found in different individuals
What is the difference between polymorphism and mutations?
polymorphism- variations that occur in more >1% of the population
mutation- variations that occur in <1% of the population
What is an SNP?
single nucleotide polymorphism
the most common DNA sequence within a population is called the ___________.
wild type