exam 2 evolutionary
1/102
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
103 Terms
What are the sources of genetic variation?
Point mutations
Recombination during crossing over
Segregation and independent assortment
Transposable elements
Chromosomal Fusion-Fission
Polyploidy
what are the various types of point mutations?
DNA synthesis and repair
Silent
Missense
Sense
Nonsense
Frameshift
SIlent Point Mutations
Not affecting primary structure of polypeptide chain
Change in nucleotide sequence that does not change the amino acid specified by a codon
Missense mutation
One nucleotide is replaced by another
Ex: Melanocortin 1 receptor
Change a nucleotide sequence that changes the amino acid specified by codon
Nonsense mutation
Change in nucleotide sequence that results in an early stop codon
Created protein that is shorter (from amino acid → stop codon)
Ex: Cystic fibrosis
Sense mutation
Stop codon is converted into amino acid → elongated protein
Hemoglobin Constant Spring → lengthening causes anemia
Frameshift mutations
Nucleotide deletions or insertions
Adding on extra nucleotide effects the frame and codon sequences
When do point mutations originate?
Point mutations originate during DNA replication or repair errors
Transitions vs. Transversion
Transitions: from one purine to another (adenine to guanine)
Transversion: from a purine to pyrimidine, vice versa
When do Trinucleotide repeats originate?
They arise during replication when DNA polymerase “slips”, expanding repeat regions. Gene expression or protein function is disrupted.
Same sequence multiple times
Premutation allele → 60-200
More than 230 → mutant allele
Which disorders are caused by Trinucleotide repeats?
Fragile X syndrome: Severe cognitive dysfunction, harms boys because they only have 1 X chromosome
Huntington’s disease: Unwanted movements, behavioral changes, paralysis, death
Myotonic dystrophy: Muscle weakness, myotonia (delayed relaxation), cataracts, cardiac arrhythmias
Which mutations originate during crossing over?
Duplication → longer segment, prophase 1, meiosis 1. A piece of the chromosome is copied and inserted, while leaving the original piece in place
Deletion
Inversion → piece of chromosome is flipped upside down, reversing its direction
Translocation → piece of chromosome moves to a different chromosome (that is unrelated), will be rearranged
Explanation of Mutations that originate during crossing over
Crossing over: 2 chromosomes line up perfectly
Mispair → overlap chromatids → some chromosomes inherit 3 copies or 1 copy (not involved in equal exchange)
How do gene families originate?
Gene duplication, combining polypeptides
Duplication creates an extra copy of a gene that accumulates mutations and evolve new functions while the original copy maintains its role
Example of a gene family
Globin gene loci in Humans and Mice
Over evolutionary time, an ancestral globin gene duplicated many times, allowed for slightly different functions
Hemoglobin different throughout life
Embryonic HB → hae greater affinity for oxygen, good for extracting O2 from mom)
Adult Hb → no more affinity
Which mutations originate during segregation?
Errors in meiosis, such as nondisjunction, unequal segregation. Lead to aneuploidy or chromosomal abnormalities (monosomy, a missing chromosome or trisomy, an extra chromosome)
Recombination and Fertilization generate variation
Meiosis: when gametes for 2 homologous that exchange → create new combination of alleles
Two distinct organisms mating, increase DNA variation
Independent assortment ensures novel combinations of alleles
Way chromosomes segregate when gametes form
more pairs, more variability in gametes (from 2 homologous pairs)
What are the various types of transposable elements?
DNA sequences that move to many places in the genome
DNA transposons → Detach from original location, insert to another place in he genome (nonreplicative), enzyme cuts and inserts into new location
Replicative DNA transposon → copied itself, inserted copy in other location of genome
Retrotransposons → Move around genome using an RNA intermediate (complementary inserting itself somewhere else in the genome)
First transcribed into RNA, reverse transcribed back into DNA by reverse transcriptase
Inserted into new loc in genome
DNA transposons
Detach from original location, insert to another place in he genome (nonreplicative), enzyme cuts and inserts into new location
Replicative DNA transposon → copied itself, inserted copy in other location of genome
RNA transposons
Move around genome using an RNA intermediate (complementary inserting itself somewhere else in the genome)
More potential to cause mutations
First transcribed into RNA, reverse transcribed back into DNA by reverse transcriptase
Inserted into new loc in genome
Ex: LINE (reverse transcriptase) → long interspersed nuclear elements, code for retro transcriptase. Autonomous, independent
SINE → short, interspersed nuclear elements
What are the effects of transposition?
Insertional mutagenesis → mutation caused by the insertion of foreign/mobile DNA into a gene, leading to disruption or misregulation of gene function
Gene disruption
Altered gene expression
Genomic rearrangements
How can we determine chromosomal fusion?
Two chromosomes fusing into 1 chromosome, where there are extra centromeres and telomeres in the middle of the chromosome
By comparing karyotypes and looking for shared telomere sequences or centromere remnants
Ex: human chromosome 2 originated by fusion of two ancestral ape chromosomes, has telomeres in the middle
How does genetic variation arise in prokaryotes?
Through polyploidy (organisms that have 3+ copies of each chromosome)
Mutation, horizontal gene transfer (transformation, transduction and conjugation)
Population genetics
study of allele frequencies and changes in allele frequencies in a population (in a particular population, genotypes)
What are the assumptions of the Hardy-Weinberg Equilibrium?
Population is infinitely large
No selection
There is no mutation (no net mutation)
Mating is random
There is no migration (no net migration → population leaving = population entering)
What are the predictions of Hardy Weinberg about allelic and genotypic frequencies?
Allelic and genotype frequencies remain constant over generations in the absence of evolutionary forces (mutation, selection, migration and drift)
Allele frequencies predict genotypic frequencies
Reject null hypothesis if we do not see what is expected by Hardy Weinberg
What is heterozygote advantage?
Heterozygotes that have higher fitness than either homozygote (overdominance)
Ex: Sickle cell heterozygotes (HbA/HbS) resist malaria infection
What accounts for the high frequency of thalssemia carriers in countries around the Mediterranean and in Southeast Asia?
Resistance to malaria
Infected red blood cells tend to sickle (more likely to rupture, cleared out by spleen more effectively)
Clears out old and nonfunctional RBC
Red blood cells produce more superoxide anion (O2-) and hydrogen peroxide (H2O2) toxic to pathogens
Reactive hydrogen species, more effectively kill pathogens
Which of the following statements is untrue about gene duplications?
They occur during DNA replication
Point mutations occur during
DNA replication
Which evolutionary mechanism can lead to adaptation?
Natural selection
Which evolutionary mechanism can increase genetic variability of a population?
Migration
Insecticide Resistance in C. Pipiens
Increase production of enzyme that breaks down insecticide
Overproduce enzyme → resistance
Pleiotropy
mutation in a single gene affects many phenotypic traits
Net effect on fitness determines outcome of selection'
Increase resistance to pesticide
Increased susceptibility to predation
Low productive success in males
Increased fitness in E.coli
Unique events occur for each population of E. Coli
Those who arrive at 50k generations exhibit improved fitness, although they do not end up at the same point
Negative selection
Alleles that lower fitness experience are eliminated
Positive selection
Alleles that increase fitness
What is genetic drift?
Random change in allele frequencies due to chance, strongest in small populations
Sampling error
When sample is too small, is not representative of population in bowl
Sample error is higher with a smaller sample
What populations are primarily affected by genetic drift?
Observed best in small populations, where some alleles are being lost and some become very common (rapid fixation)
Ex: Genetic drift in D. Melanogaster
over time, more and more populations lose a allele and become fixed for either bw or bw75
bw75 was lost, both reached fixation
What is the bottleneck effect?
A population undergoes a drastic reduction in size; survivors’ alleles dominate future generations
Ex: Northern elephant seal
Subject to hunting
N = 30 survivors, mtDNA, 2 polymorphisms (2 specific loci in the same stretch of DNA)
Reduce number of individuals that reproduce
Allele loss probability in Bottlenecks
p = 0.01 → allele is rare, more likely to be lost
p = 0.10 → common
Smaller population, more probability on allele will be lost
Haplotype
Several genetic markers located on same chromosome
Population can have several haplotypes
Polymorphism
Specific locus in same stretch of DNA
Variation in one locus
What is founder’s effect?
When a few individuals colonize a new area, they carry only a fraction of total genetic variation
Colonizing a new environment (areas that are isolated and lack their species
New populations started by a small number of individuals
Founder effect causes genetic drift
Ex: Black spruce expansion, forest expands (more north reduced)
Founder effect → Mutiny on the Bounty
Background: British ship crew rebelled against captain, took captain and pushed out to sea
Lead to the colonization of Pitcairn (1789)
→ 11 mutineers, 6 Tahitian women
As population increased, they moved to a new island, Norfolk (1854)
193 colonists → population 2,000 (today)
Which evolutionary sources lead to adaptation?
Natural selection and mutation (raw variation)
What are the effects of genetic drift and gene flow?
Drift: decreases variation, causes random fixation/loss of alleles
Gene flow: increases genetic variation, but disrupts local adaptability
Causes allele frequencies to be more alike (in two populations)
What are the differences between Mendelian vs. quantitative traits?
Mendelian traits: monogenic or discrete (single gene)
Quantitative traits: continuous, polygenic traits influenced by environment (influenced by many genetic loci, 4 genes for 1 trait)
What are multifactorial traits?
Heritability (construct that estimates genetic variance to environmental variance)
Traits influenced by multiple genes and environmental factors
What is heritability?
Statistical construct that estimates the amount of variation that is attributed to genes
Applies to a specific population in a specific environment
Explains the variance among individuals, not the character itself
Ex: height heritability
Example of heritability
Ex: Height heritability
0.8 → variance b/c of genetic makeup (additive effects of recessive alleles, dominant alleles, epistasis (one gene influencing expression of another gene)
0.2 → b/c of environmental factors
What is absolute genetic determinism?
Traits entirely determined by genotype with no environmental influence (Ex: blood type → only genes that affect the blood type)
How does Broad Sense heritability differ from Narrow Sense Heritability?
Broad sense (H²): includes all genetic variance (additive, dominance, interaction): H² = VG / (VG+ VE)
Narrow sense (h²): includes ONLY additive variance → h² = VA / (VG + VE)
Broad sense (H²) equation
H² = VG / (VG+ VE)
Narrow sense (h²) equation
h² = VA / (VG + VE)
Breeder’s equation
R = h² x S
R = response to selection
h² = heritability
S = selection differential
What are the various types of natural selection?
Directional
Stabilizing
Disruptive
Balancing
Directional selection
One extreme phenotype has advantage (selected for)
Ex: Elephant tusklessness (females)
In early 70s → only 18.5% of females were missing tusks
1977-1992 → War rebels killed elephants and sold tusks for armor
Strong selective pressure, one extreme was selected → 50.9% missing tusks
Number (as of most recent) has shifted to 58.4% having tusks
Stabilizing selection
Average phenotype has advantage
Example: Aposematic coloration stability
Warning color for predators (prey is nonpalatable)
Tested reflectiveness of yellow and red on body as there is variation in spots and color
Limited variation between sites
Some variation (reflective on red), limited variation (reflective of yellow)
Constant average was associating yellow with unpalatable food (predators)
Disruptive selection
Two extremes → advantage
Average → disadvantage
Pushing population to 2 extremes
Bimodel distribution
Example: Color morphs in stick insects
Example of disruptive selection in stick insects
Polymorphic (T. Chumash), also has intermediate and melanic → greater variability in host plant colors
T. Bartmani → have green and brown → melanic morphs (match host plant leave and twig colors)
T. Cristinae → green and brown - mostly match green leaves and brown twigs
Mark insects, see how many return after capture (T. Chumash)
40 of each in each treatment (green, intermediate and melanic)
In treatment with host plants of other 2 insects, two species show disruptive selection (2 extremes)
Balancing selection
Multiple alleles are being maintained (polymorphism) → different alleles
Heterozygote advantage → sickle cell anemia, at an advantage where malaria is
Negative frequency dependent selection → when allele is rare, selected for (higher fitness)
Fluctuating selection → when rare allele is selected, it becomes common, common allele is blocked
Example of fluctuating selection (T. cristinae)/Balancing selection
Three morphs of unstriped, striped, melanistic
one morph: green, striped (frequency is high) → becomes low → returns high
Visual predators → create search image for high frequency morph (rare) → becomes common, predators hunt for this search image
Strong vs. Weak selection for Body size
Usually goes for strength of selection → area between mean size of entire pop. and mean size of reproducing individuals
h² = 0
Heritability of 0 → not passed on, no genetic contribution
population will remain consistent
R → response
h² = 1
R = S → response equals strength of selection
Which factors affect the degree to which a population will evolve?
Heritability
Strength of selection
Mutation
Gene flow
Drift
What is linkage disequilibrium?
Alleles on same chromosome
Loci that are adjacent to each other are tightly physically linked, inherited together
How do genetic markers work?
BRCA1 gene → tumor suppressor
Linkage disequilibrium with all loci along the entire chromosome → mutation of BRCA1 increases risk of breast cancer → disables gene
Recombination occurs → crossing over during meiosis
After 4 generations, linkage disequilibrium remains (decreases in size), SNPs tightly physically linked with a disease risk allele can be used as a genetic marker for the presence of this allele
What are SNPs?
Single Nucleotide polymorphisms
Linked SNPs
outside of gene → no effect on protein production or function → can indicate where gene is located
non coding SNPs
changes amount of protein produced
coding SNP
changes amino acid sequence
Causative SNPs
in gene
non coding SNP → changes amount of protein produced
Coding SNP → changes amino acid sequence
What are Variable Number Tandem Repeats?
Variable number tandem repeats (DNA sequences)
varies for each individual
basis for genetic fingerprinting
What are the various applications of LD?
Estimate mutation origin → size of link. diseq→ big in new population, small in old population
QTL analysis
GWAS→ genes under selection, selection effects, gene identification
Indicator of positive selection
Test historical hypotheses → Ex: Did Polynesian travel to South America? → took ancestral DNA and modern DNA → short around SA markers (recent), large with ancestral DNA
How is QTL analysis done?
Statistical association of genetic markers with phenotypic variation in controlled crosses; typicaly uses LOD ≥ 3 as significant
Breed largest individuals tg. And smallest individuals to create 2 homozygous
Generations will be heterozygous
Observe high likelihood ratio (marker and trait association)
Where genes related to characteristics are used → if parallel, they are inherited tg.
QTL analysis of coat color in mice
Beach mouse
Mainland
1st gen hybrid → created
2nd gen → heterozygous for every loci
Some will be light, some will be dark
F2 = 465 → polygenic mutation , multiple genes
Interaction of Agouti and MC1r
Agouti (gene) → Mc1r inhibits (if there is a missense)
If there is high signaling, dark pigment is expressed
Low signaling (has a missense), no pigment is expressed
MSH (melanocyte stimulating hormone) → bonds to receptor (Mc1r)
Epistasis
Interaction between genes where one gene masks or modifies the effect of another on the phenotype (can effect trait heritability and selection responses)
Mice → DD at the agouti locus, Mc1r has no effect on the phenotype
DL OR LL → at Agouti locus, the alleles at the Mc1r locus influence phenotype (important for coloration)
Which alleles are involved in phenotypic expression?
More dark pigment on the dorsal (back) → more dark fur
No pigment on ventral belly → less dark pigment expressed → bicolor beach mice, greater degree of Agouti
Precise mechanism of Agouti
Is responsible for maturation of melanin
Delays melanocyte maturation
Strongest ventral expression (side of belly)
Non-Agouti peromyscus
Experienced gene deletion, cannot be expressed
Is fully dark, hair follicle has no color because of Agouti (when expressed) → inhibits color of melanin (have MLV + Agouti)
Control → no working Agouti → melanocytes developed hormonally (just MLV)
Genetically manipulated dark mice
Are homozygous for Agouti allele LL
Shows ratio of expression
Intermediate from dorsal midline
What is the significance level in QTL studies?
LOD ≥ 3 as significant
What are GWAS?
Genome Wide Association Studies
Genetic markers for diseases (SNP-trait associations in populations)
Looking at entire genome → limited to organisms where several genomes have been sequenced (Model organisms)
Ex: patients with coronary artery disease and those who do not
p < 5 × 10^8 = -7.3
-log (0.00000005) = 7.3 → any marker under 7.3 is insignificant
What is the significance level in GWAS studies?
p < 5 × 10^-8
What is phenotypic plasticity?
Expression of gene influenced by environmental factors
1 genotype can generate many phenotypes in environment
Shows norm of reaction
Ex: low light vs high light
Example of plastic plasticity 1
Low biomass with low light environment, bigger leaves are produced (photosynthesis need increases due to lack of light) → produces more chlorofilm
High biomass with high light environment (smaller leaves, receives sufficient amount of photosynthesis)
Norm of reaction
Set of phenotypes produced by genotype
What is Polyphenism?
Discrete environmentally induced phenotypes
Ex: How many nutrients are present?
On beetles, low nutrients causes no horns
High nutrients, male has horns
How can we detect genotype environment interactions?
By measuring phenotypic responses across multiple environments and comparing reaction norms
What is gene coalescence?
Tracing alleles in a population back to a common ancestral copy (most recent common ancestor)
How can gene trees differ from species trees?
Gene trees trace the ancestry of a particular gene
Species trees trace the ancestry of an entire species
Incomplete lineage sorting or gene duplication → causes gene trees to not always match species trees
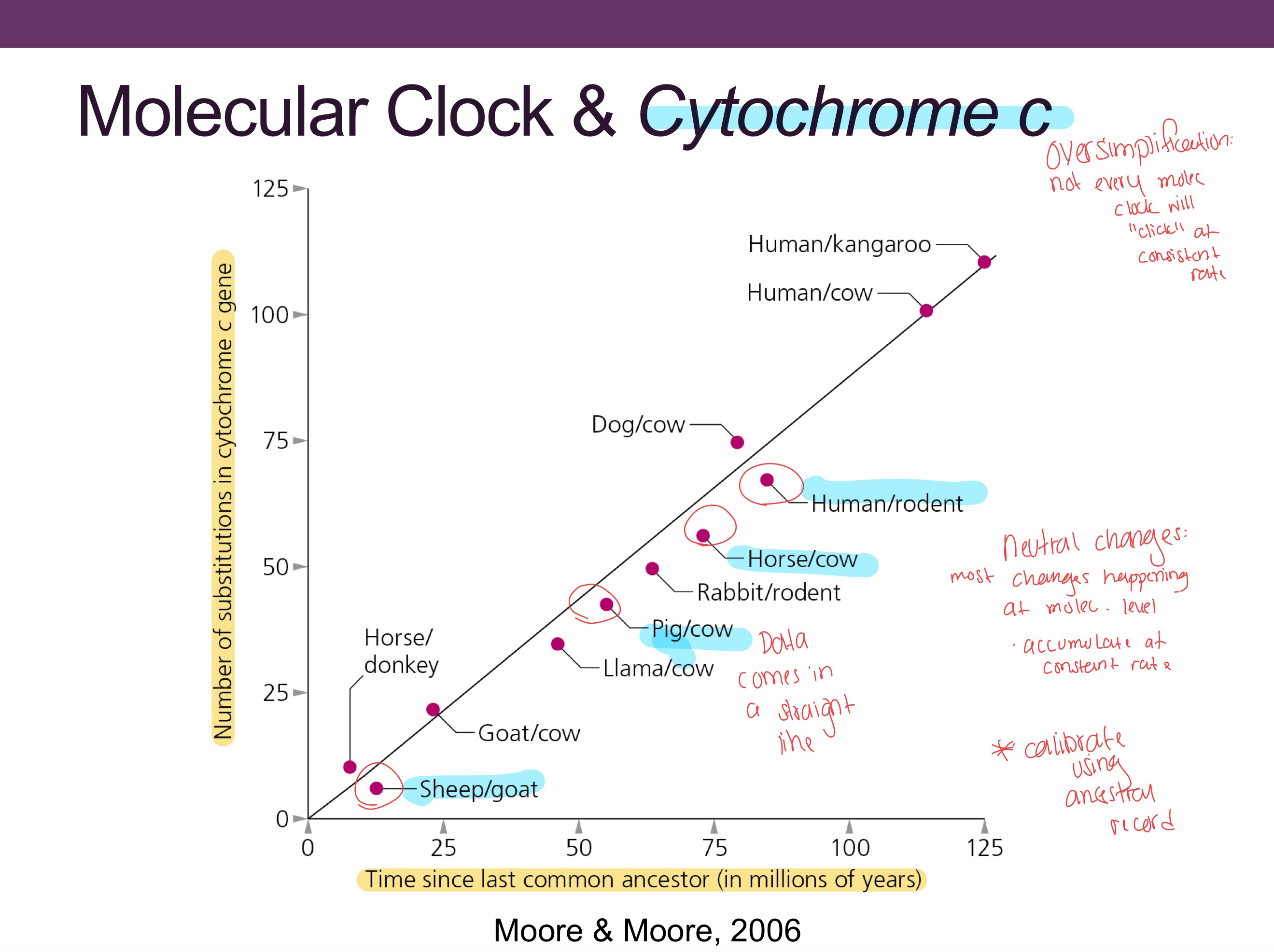
What is a molecular clock?
Assumes mutations accumulate at a roughly constant rate, allowing estimation of divergence times
Which areas of the genome evolve faster/slower?
Faster: noncoding, pseudogenes, synonymous sites
Slower: essential genes under strong functional constraint
What is Maximum Parsimony?
Tree with fewest character changes
Creating phylogenetic tree with lowest number of changes
What is bootstrapping and how is it used in the construction of phylogenetic trees?
Bootstrapping: testing tree robustness using character subsets. Statistical resampling of data to test the robustness of tree nodes. Higher bootstrap values indicate stronger support
Originally 4 species, 10 characters
Using those with the highest confidence (100)
What is the method of neighbor joining?
Joining species that have the fewest differences together (clustering taxa by minimizing total branch lengths)