5. modes of inheritance I
1/46
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
47 Terms
Species
group of organisms comprised of similar individuals capable of interbreeding
Population
individuals of a particular species occupying a definite space, in which individuals interact, interbreed, and exchange genetic material
Gene pool
all alleles from all individuals within a population
Allele/ genotype frequency
proportion of individuals in a population with a specific allele/ genotype
may differ among populations
Population genetics
Quantitative study of the distribution of allele frequencies in a population and its changes over time and between populations
Mathematical modeling of allele behavior offers insight into the genetic susceptibility to disease
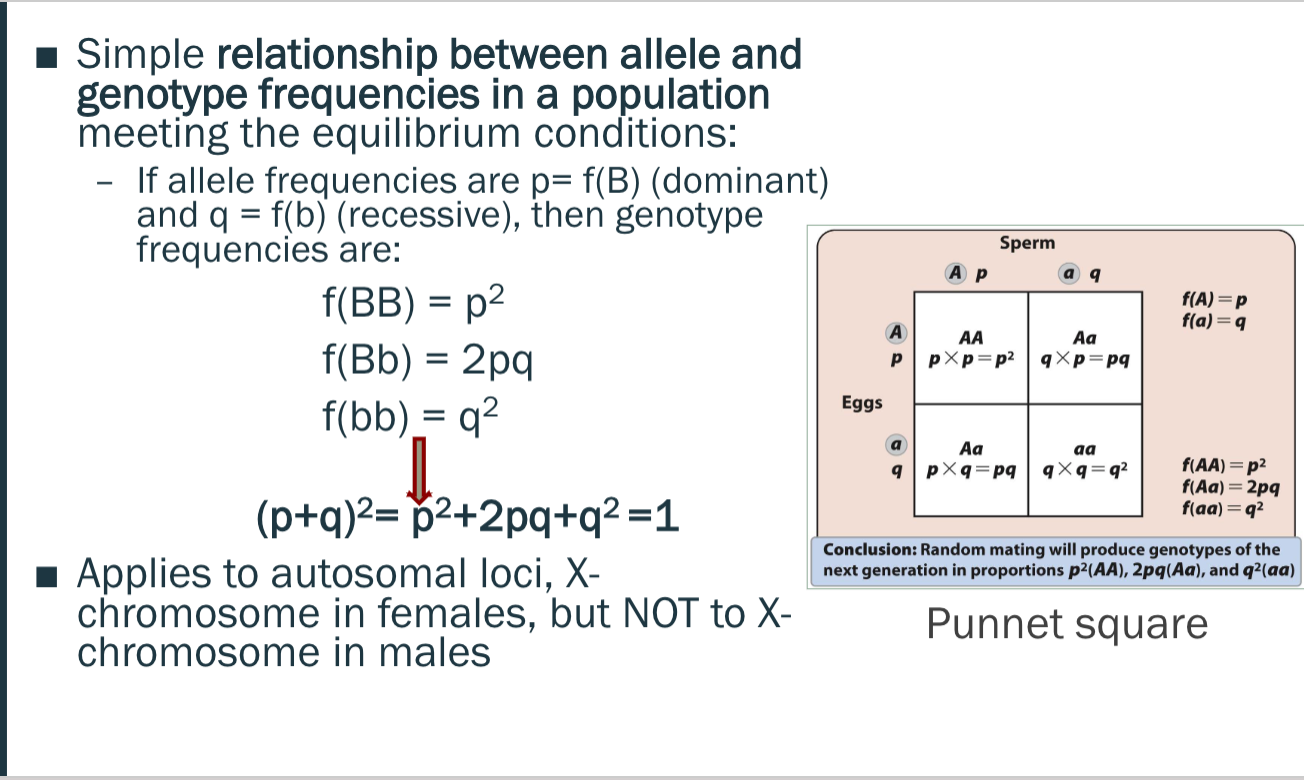
Hardy-Weinberg law
Allele frequencies remain constant over time in a population that does NOT evolve = genetic equilibrium
allele frequency calculations
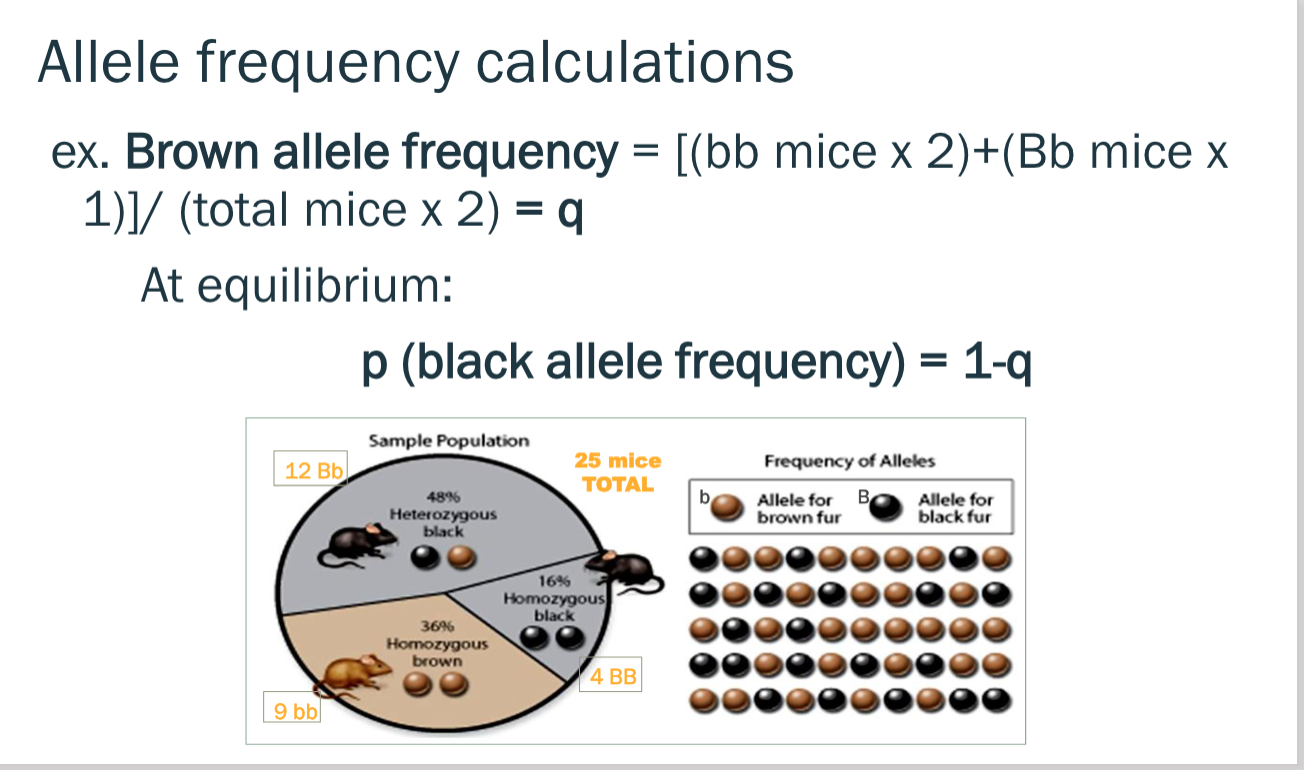
factors that disturb hardy weinberg equilibrium
1. Non-random mating
2. Small population size
3. Mutations
4. Natural selection
5. Immigration and emigration
Small population size
random effects (survival, fertility), unrelated to carrying the mutant allele, can
change allele frequency between generations
Mutations
new alleles introduced in the gene pool
Natural selection
predilection for a positive allele → disruption of equilibrium
Immigration and emigration
addition or removal of alleles
Non-random mating
Increased frequency with which carriers mate → substantial deviation from equilibrium
Stratification
population with subgroups that remained genetically separate
If that subgroup has an allele with higher frequency than of that the whole groups → apparent excess of homozygotes in the overall population
No effect on frequency of autosomal dominant disease
Assortative mating
Choice of mate based on a particular trait
Long-term effect is minor
Consanguinity and inbreeding
Recessive disorders in offspring of related parents are rare and unusual
(eg. Tay-Sachs disease in the Ashkenazi Jew population 1:3600 vs. 1:360000 in general population)
Mutations and selection
Occur very slowly and in small increments inducing small deviations from the HW equilibrium
Most detrimental recessive alleles are hidden in heterozygotes
selection has no short-term effect
Fitness (f)
measure of surviving affected offspring compared to control (outcome of collaboration between survival and fertility)
Main factor that determines whether a mutation is lost, becomes stable, or even becomes dominant in time
f=1 means
a mutant allele is as likely as the wild-type to appear in the next generation
f=0 means
the allele causes death or sterility or is negatively selected against
Stable allele frequency
balance between removal (selection) and addition (mutations) of mutant alleles to a gene pool
Gene flow
slow diffusion of genes through barriers
Involves large populations and slow change in allele frequency
Genes of migrant populations are slowly merging in the gene pool of
the population they migrate into (similar neighboring populations)
ex. DCCR5 mutation – highest frequency in Europe, small in Middle
East and India and almost absent in Africa
Ethnic differences in allele frequency
Until recently, humans lived in isolated groups = larger differences in allele frequency between populations
Factors that influence differences in alleles and allele frequency among ethnic groups
genetic drift and heterozygote advantage
Genetic drift
random change in allele frequency in small populations due to chance
Individuals with the mutant allele may have more offspring (by chance) → allele becomes frequent in population in time
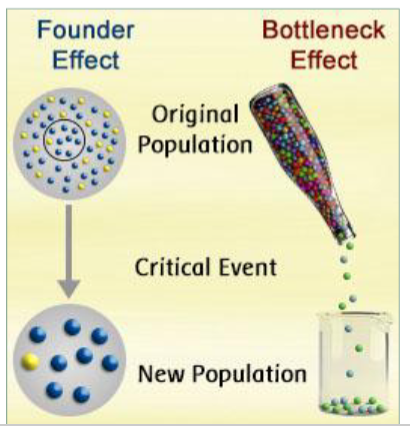
founder effect & bottleneck event
Founder effect
genetic drift after a few individuals (a fraction of the pool) start a new population in isolation from original population (different frequencies)
Bottleneck effect
genetic drift after event that drastically reduces population size (might have different allele and genotype frequencies)
Heterozygote advantage
when heterozygotes have increased fitness over wild-type homozygotes
Increased frequency of mutant allele
ex. Sickle cell anemia and malaria
Changes in selective pressure → change in frequencies
Single-gene disorders (mendelian)
Determined primarily by alleles on a single locus
Follow primarily one of the classical inheritance patterns (autosomal dominant/recessive, X-linked dominant/ recessive)
OMIM – approx. 8000 single-gene disease or traits
in humans
Pleiotropy
a single abnormal gene → a variety of phenotypes in different organs, with different signs and symptoms, at different times
Penetrance
probability that a mutant alleles will have a phenotypic expression (all-or-none)
If a genotype fails to express = reduced/incomplete penetrance
Age-dependent for some disorders
Expressivity
degree of phenotype severity
variable expressivity
Severity of symptoms is different in individuals with same genotype
pedigrees
Graphical representation with standard symbols of a family tree to establish the pattern of transmission
proband (propositus/a or index case)
First individual diagnosed with disease in a family
how to interpret pedigrees: 1. Autosomal vs. sex-linked
Mostly males → likely X-linked
50:50 M:F → autosomal
Male-to-male transmission → autosomal
how to interpret pedigrees: 2. Dominant vs. recessive
One parent must have the disorder → dominant (2 affected parents can have an unaffected child)
Neither parent must have it → recessive (2 unaffected parents can have an affected child)
Autosomal dominant inheritance (AD)
Only one copy of a diseased allele is required for the possibility to exhibit the phenotype
Quite rare, but >50% of all mendelian disorders
Eg. Huntington’s disease, polycystic kidney disease, familial hypercholesterolemia
characteristics of AD
No skipping generations - each individual has at least one affected parent (unless it is a new mutation)
The recurrence risk for each child of an affected heterozygote parent is 50%
A significant proportion of cases are sporadic, due to de novo mutations
Both sexes display the phenotype in equal ratios and are as likely to transmit it to their offspring
Normal siblings of affected individuals are not carriers of the disease
Defective gene product is usually a structural protein, transcription factor
Autosomal recessive inheritance (AR)
Phenotype exhibited only when both alleles are mutated
Eg. albinism, phenylketonuria, alkaptonuria, sickle cell anemia, cystic fibrosis
Characteristics of AR
Males and females are equally affected
The recurrence risk for the offspring of 2 heterozygotes is 25%; if a homozygote mates with a heterozygote, the recurrence risk is 50%
Multiple siblings in the same generation manifest the trait (not in parents or offspring)
Parents of affected individuals are generally asymptomatic carriers and in some cases may be related (skips generations)
Products of affected genes are mostly enzymes
Y-linked inheritance
Very few genes (Y-specific related to spermatogenesis or primary determination and some housekeeping with X- homologs)
Transmitted strictly from father to son in all generations
Affects ONLY males
X-linked inheritance
Easy to distinguish from autosomal inheritance
Possible genotypes:
Male → hemizygote
Female → homo/heterozygote
X-linked recessive inheritance
X-linked loci are similar to autosomal loci for females; however, due to randomized X-inactivation, half of cells will express the normal allele and half the mutated one in heterozygotes
Eg. hemophilia A, Duchenne muscular dystrophy, colorblindness
Characteristics of X-linked recessive
Higher incidence in males than females
Heterozygous females generally unaffected
Trait skips generations
Mutant allele never transmitted from father to son!
All daughters of affected fathers are carriers!
X-linked dominant inheritance
Very few disorders and less prevalent
Expressed even in heterozygotes (affects both females and males), similar to autosomal dominant
Characteristics of X-linked dominant inheritance
Distinguished from autosomal dominant by lack of male-to-male transmission
Affected males with normal mates → no affected sons and no normal daughters
Affected females mating with normal males produce ½ sons and ½ daughters affected
Males have usually a more severe phenotype (might be lethal)
Affected females are twice as common as affected males