Chapter 11 - DNA Replication (Part 2)
1/14
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
15 Terms
DNA Polymerase III
The enzyme that replicates the E.coli chromosome. It is a large multisubunit protein
Core Enzyme
The number of subunits that are capable of the base function of the enzyme.
Holoenzyme
When a core enzyme is associated with additional components to enhance function.
1) Subunit with polymerase activity
2) Subunit with 3’ to 5’ exonuclease activity (proofreading)
3) Same hand shape as other polymerases
What are the components of the polymerase III core enzyme?
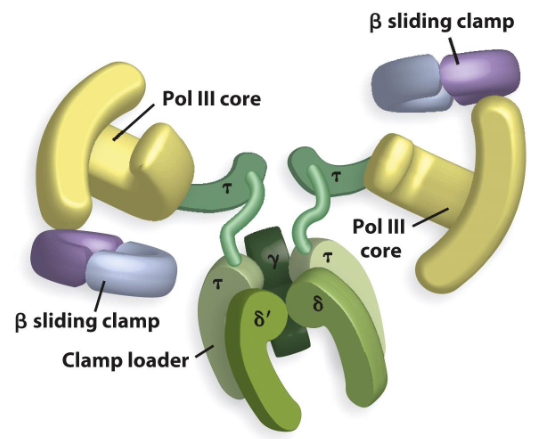
1) Two beta sliding clamps
2) Clamp loader
What are two components not in the core enzyme but are in the holoenzyme of polymerase III?
Slow / holoenzyme
Fill in the blank…
The core enzyme of polymerase III can synthesize DNA at a ( ) rate. The ( ) is capable of making DNA at about 1 kb/sec.
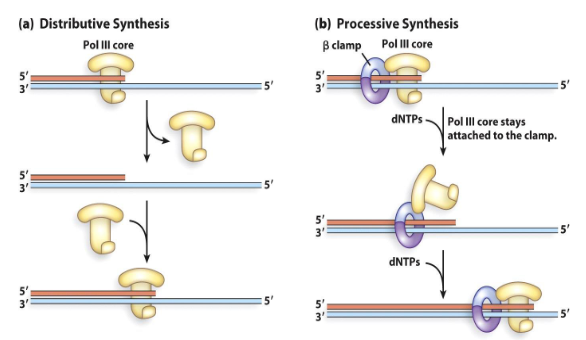
Sliding Clamps (Beta)
These components of the holoenzyme help the DNA polymerase to be processive. It keeps the DNA polymerase from diffusing away from the primer template junction.
It stays on DNA for more than 100 kb per binding event.
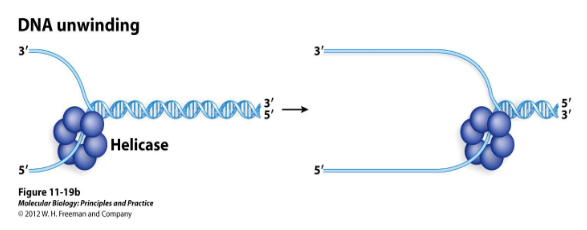
Helicase
A protein that opens up double-stranded DNA to expose the bases by using the energy of ATP. It moves in only one direction.
Topoisomerases
Proteins that relax the positive supercoiling in DNA before it breaks. This overwinding is often introduced during the action of helicase. This is most needed for circular molecules, but it can occur in linear molecules.
Single-Stranded Binding Protein
A protein that coats the single-stranded DNA following separation by helicase and keeps it single-stranded. It binds cooperatively and is sequence independent. It has no bonds binding it to the DNA; it uses electrostatic and hydrophobic interactions.
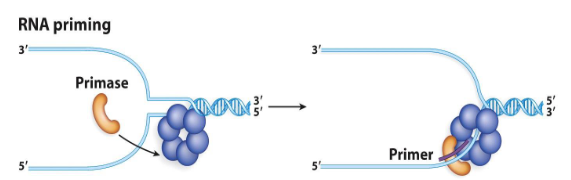
Primers
RNA sequences that provide the first 3’-OH group for replication. No DNA polymerase can start replication without this. Cells will take advantage of RNA polymerases to make RNA and allow production of short RNA sequences that initiate synthesis of Okazaki fragments on the lagging strand.
These sequences are usually 11-13 bases long.
Primases
Specialized enzymes that make primers. They are different from RNA polymerase in that they do not require a specific DNA sequence to get started. They are only active when the unwound DNA is still in association with the helicase; this localizes their function to the replication fork.
Replisome
The combination of all the proteins at the replication fork.
1) Initiator protein binds to the origin of replication
2) Helicase unwinds the helix
3) Single-stranded binding proteins keep the DNA helix open
What occurs in initiation stage #1?
1) Primase synthesizes RNA primers that are complementary and antiparallel to each template strand
What occurs in initiation stage #2?