bio 108 quiz 3
1/139
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
140 Terms
what are monomers of nucleic acids
nucleotides
what sugar does RNA and DNA have.
RNA has ribose
DNA has deoxyribose
what is the structural difference between ribose and deoxyribose
ribose has an OH bond at C2’
deoxyribose has an H bond at C2’
can RNA be double stranded? what is the structure known as
yes, RNA can have double stranded regions which are self complementary
this is known as a stem loop
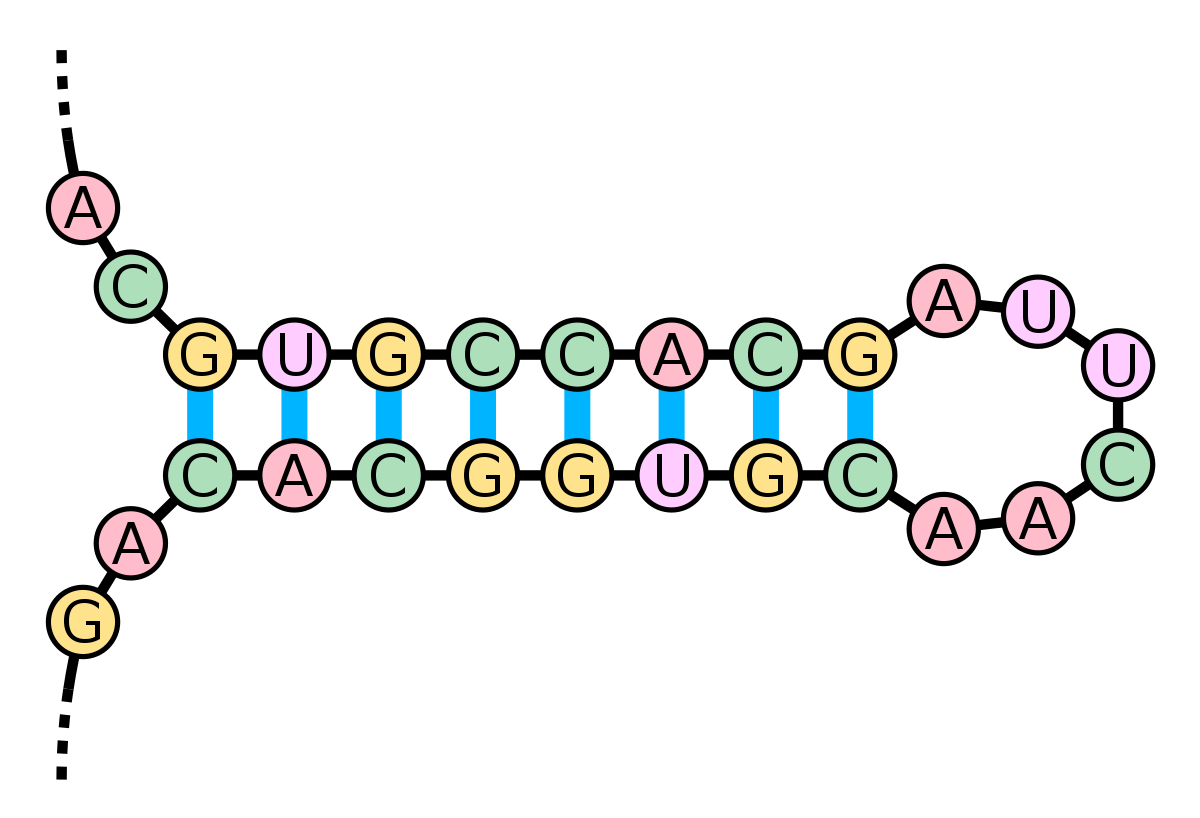
what is need to form a stem loop
you need all the 4 bases to form a stem loop
describe the strands of DNA
DNA is double stranded and antiparallel
what are the DNA nucleobases
Guanine (G)
Adenine (A)
Thymine (T)
Cytosine (C)
what are the RNA nucleobases
Guanine (G)
Adenine (A)
Uracil (U)
Cytosine (C)
what are pyrimidines? what bases are considered pyrimidines
pyrimidines refers to bases with single ring structures
Cytosine, Thymine, and Uracil are considered pyrimidines
what are purines? what bases are considered purines
purines refers to bases with double ring structures
Adenine and Guanine
what are the components that made up a nucleotide
base + ribose or deoxyribose + phosphate
what are the components that made up a nucleoside
base + ribose or deoxyribose
what is the difference between thymine and uracil
at C5, thymine has a methanol group,
at C5, uracil has a hydrogen group
describe the complementary base pairing in DNA
A - T
G - C
describe the complementary base pairing in RNA
A - U
G - C
describe how complementary base pairing occurs
purines pair with pyrimidines by hydrogen bonding
which base pair is harder to separate?
G-C base pairs have 3 H-bonds which are harder to separate than A-U or A-T base pairs which have 2 H-bonds
describe the regions of DNA where a separation happens
these regions are A-T rich because A-T is easier to separate than G-C
what does two DNA strands form
they form a double helix
all DNAS
what is the name of the strand that is synthesized during DNA Replication
its called the primer strand
what is the name of the strand that is copied during DNA replication
its the template strand
how does the primer strand elongate during DNA replication
by the addition of DNA precursor molecules called deoxynucleotide triphosphate
what falls under deoxynucleotide triphosphate? what are they referred to as?
they are referred to as dNTPs
dATP,
dCTP,
dGTP
dTTP
what does the “d” stand for in dNTPs
it stands for deoxyribose
what does the “N” stand for in dNTPs
N can stand for A, C, T, or G.
describe the reaction and the mechanisms involved in dna replication
the 3’ end of the primer strand has an OH group
the oxygen (O) from that hydroxyl group attacks the alpha phosphate of a DNA precursor
what is the name of the direction of DNA synthesis
it is called 5’ to 3’
this is because 3’ side is where the new bases are continuously being added
what length is getting longer in the primer strand
the 3’
what happens to a dNAP during DNA synthesis
the alpha phosphate breaks from dNAP, releasing 2 phosphates that are linked together, called pyrophosphate
a new bond is formed called a phosphoester bond
what does DNA synthesis begin with?
DNA synthesis begins with short pieces of RNA (10-15 nucleotides)
what must occur in order to convert two strands of DNA into four
DNA synthesis must occur on both strands of DNA in order to convert two strands into four
what is a replication fork
the site where both strands of DNA are being replicated
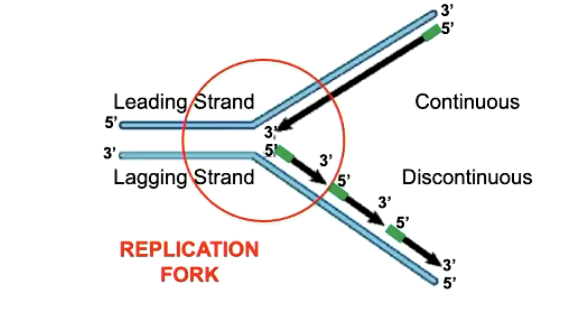
describe what happens with the top strand in a replication fork. what is the name given to it
it synthesize in a continuous way. this is known as a leading strand
describe what happens with the bottom strand in a replication fork. what is the name given to it
it synthesizes in a discontinuous way (in fragments). this is known as a lagging strand
what is the name given to the fragments seen in discontinuous DNA synthesis
okazaki fragments
what causes the difference between continuous and discontinuous DNA synthesis
because DNA synthesis only occurs in 5’ to 3’ direction
continuous DNA synthesis already moves from 5’ to 3’
discontinuous DNA synthesis occurs in lagging strands because its moving in the wrong direction relative to the direction of the replication fork.
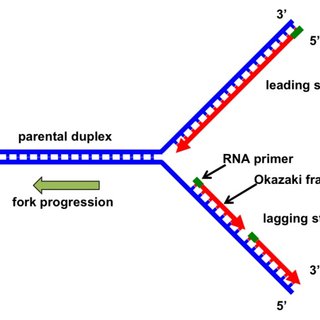
what is the site where DNA synthesis begins known as
its known as replication origin
how does replication begin and proceed? what does this lead to
it begins and proceeds bidirectionally (both ways)
this leads to two replication forks
what is the first protein added to a DNAP replication fork? what does it do
its called DNA helicase
it separates the two strands by surrounding itself on one strand and moves down.
what does DNA helicase creates while separating the strands
it creates knots as its is separating the strands
what is the an additional protein added to a DNAP replication fork after the DNA helicase? what does it do
its called DNA topoisomerase
it removes the knots created by the DNA helicase
is DNA topoisomerase in the replication fork
no its not directly associated with the replication fork.
what does the term SSB refer too? what do they do?
it refers too Single Strand (DNA) Binding proteins
it protects and organizes single stranded DNA
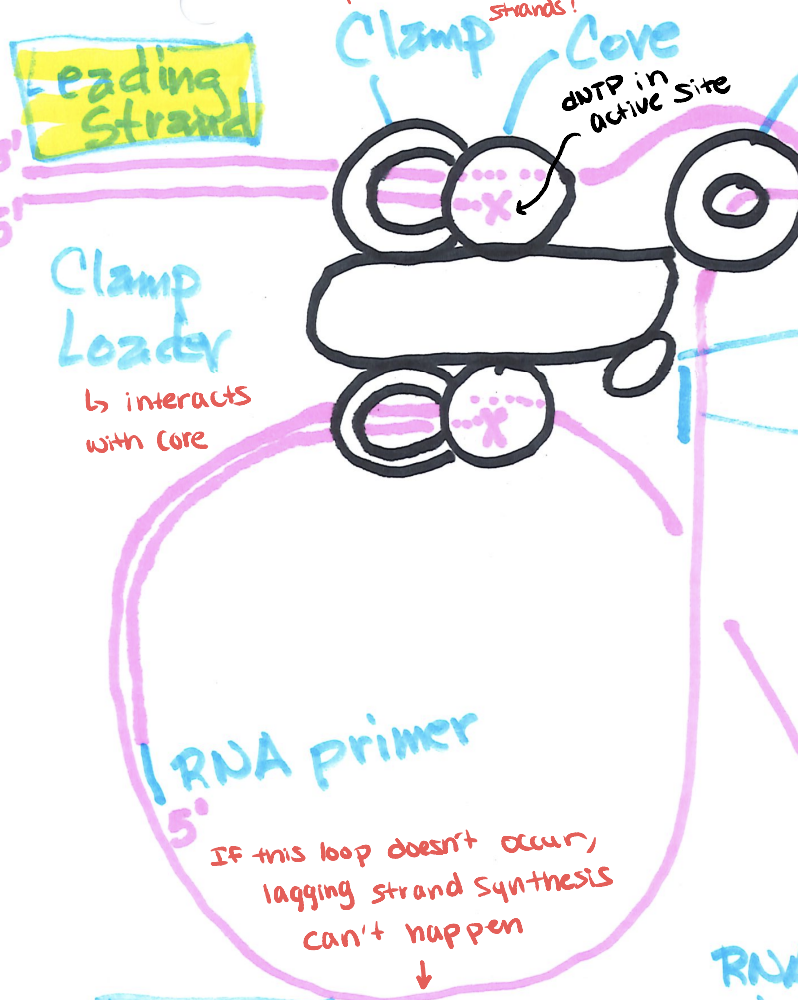
what are the proteins involved in leading and lagging strand synthesis
DNAP III- Core
Clamp
Clamp Loader
DNA Primase
what does DNAP III - Core do?
it does DNA synthesis and has 3 peptides
what does the clamp do
surrounds DNA strands and prevents DNAP from dissociating
donut shaped and is behind core
what does the clamp loader do
adds and removes clamps and DNAP III Core from double-stranded DNA when appropriate
interacts with core
has 5 peptides
what does DNA primase do
it synthesize the RNA primers needed to start DNA syhthesis
describe the coupling between leading and lagging strands
only 1 clamp loader which intersects with clamp and core complexes on both DNA strands.
what proteins although involved in the synthesis aren’t in the fork
SSB and Topoisomerase
what need to occur for lagging strand synthesis to happen
a loop
what term is referred to all of the DNA in an organism
genome
what synthesizes RNA
RNA polymerase (RNAP)
describe transcription
a process by which RNA polymerase (RNAP) copies one of the two strands of DNA to make RNA
what type of RNAs are made during transcription?
all RNAs are made during transcription by a RNAP, some include
messenger RNA (mRNA)
ribosomal RNA (rRNA)
transfer RNA (tRNA)
Noncoding RNA (ncRNA)
what is the only RNA, that is translated into proteins?
mRNA
describe translation
a process by which mRNA encodes a sequence that a ribosome copies to make a protein
what does the term central dogma refer to?
a term used to describe the two steps transcription and translation, but this term is arcane, and few people use it now.
how many types of RNAP are in bacteria
one
how many types RNAP are in eukaryotes
three
what does RNAP 1 transcribe
rRNA
what does RNAP 2 transcribe
mRNA
what does RNAP 3 transcribe
tRNA
describe the mechanisms of RNAP
its the same idea as a DNAP
the oxygen (O) from that hydroxyl group attacks the alpha phosphate of an RNA precursor
a phosphoester bond also forms
describe the direction of RNAP during RNA synthesis
5’ to 3’ - same as DNAP
what is one notable differnce between RNAP and DNAP
RNAP does not require a primer, while DNAP does.
what are the RNA precursors
ATP
CTP
GTP
UTP
is the RNA precursor ATP, the same as the one used in high energy reaction
yes
what is the name of DNA strand being copied during RNA synthesis
transcribed strand
what is the name of DNA strand not being copied during RNA synthesis
non-transcribed strand
which of the DNA strands does the newly created RNA sequence correspond to in RNA synthesis
it corresponds to the sequence of the non-transcribed strand (except the T is replaced by a U)
this is because RNA precursors are inserted opposite of the DNA strand by base pairing
describe RNAP in all species
it is similar in all species
what are the three phases of transcription
initiation
elongation
termination
what is RNAP aided by during the initiation phase of transcription
RNAP is aided by auxiliary transcription factors which are proteins
often abbreviated “TF”
describe the process of transcription in bacteria
RNAP has 6 subunits (polypeptides)
one of the subunits is called sigma
sigma recognizes 2 specific DNA sequences near TSS (transcription start site)
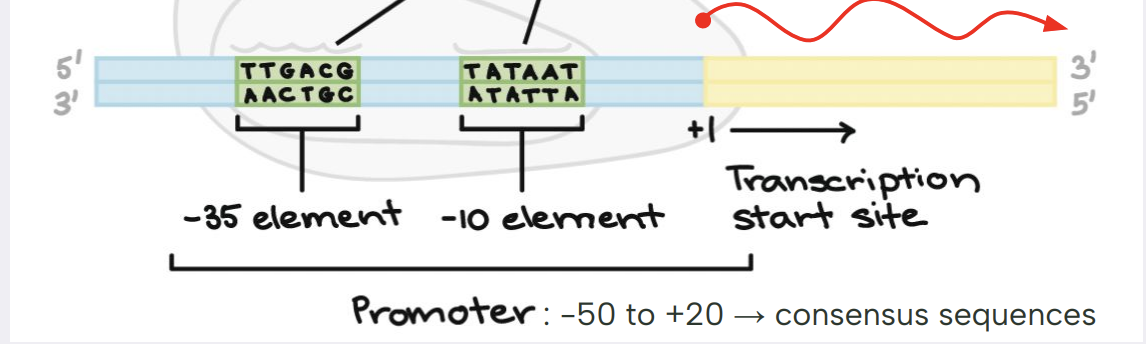
at what position is the transcription start site (TSS) in bacteria
+1
what are the consensus sequences and what is their importance during transcription in bacteria.
the consensus sequences are 6 base pairs that are recognized by sigma. they occur at two specific DNA regions
-35 sequence
-10 sequence
what does the term promoter refer to in transcription in bacteria
it refers to the RNAP binding sites:
-50 to +20
it includes the consensus sequence
what does sigma also do during transcription in bacteria
it also separates DNA strands
is the position of TSS the same in eurkayotes
yes, it occurs +1 position
describe the process of trancription in eukaryotes
there is no RNAP subunit that recognizes the transcription start site (TSS)
RNAP interacts with general transcription factors “GTFs” that bind around TSS

what are two examples of GTFs and where do they bind in eukaryotes
TBP: its binds at -30
TF(2)B: it binds at -35 and -25
upstream and downstream of TBD
what do GTFs form in eukaryotes
GTFs form a huge complex of approx a 100 peptides
what are DNA strands at TSS separated by in eukaryotes?
separated by GTFs: TF(2)B and TF(2)H
what is the eukaryote promoter definition at BU?
its goes from - 150 to +40
corresponds to where nucleosomes must be removed to allow GTFs and RNAP to bind
what are nucleosomes
protein complex with 8 histone proteins
DNA is wrapped around histones in eukaryotic celles
which nucleosome is removed?
-1 nucleosome is removed where GTF and RNAP bind at -150 to +40
what is the purpose of nucleosomes
organize and condense DNA
human diploid DNA is 2ft long and must fit into the nucleus
what controls when transcription starts?
two types of transcription factors (TFs) control when transcription starts
activators
repressors
describe the role of activators
it has a positive role
TF (aka activator) binds with DNA and recruits RNAP, leading to transcription to occur
describe the role of repressors
it has a negative role
TF (aka repressor) is bound to DNA and prevents RNAP from binding. when TF is released, that’s when RNAP binds
what is term is applied when no transcription is occurring
OFF
what is term is applied when transcription is occurring
ON
using the example of lactose metabolism, describe what happens when a repressor is bound to the DNA
no transcription is occurring
there is no lactose inside or outside the cell
using the example of lactose metabolism, describe what happens when a activators is bound to the DNA
transcription is occuring
lactose outside the cell
no glucose oustide the cell
what does having no lactose outside a cell correlate to in terms of transcription in lactose metabolism
its means that it is repressor bound
what does having lactose outside a cell correlate to in terms of transcription in lactose metabolism
its means that the repressor is released
what does having no glucose outside a cell correlate to in terms of transcription in lactose metabolism
it means that the activator is binding
what are two instances when transcription is highest in lactose metabolism
when repressor protein is not bound because it senses high lactose levels
when activator proteins are bound because it senses low glucose levels
what are the two regions in a gene in a eurkaryote
regulatory region
transcribed region