Genomics final exam
1/114
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
115 Terms
Sustained in situ protein production
and release in the mammalian gut
by an engineered bacteriophage:
What is the point of this experiment
To use phage infected resident gut bacteria to deliver therapeutic agents into the GI tract without having to take daily medication
Hypothesis of the experiment
the lytic phage T4 could help release the GFP from inside the bacteria into the surrounding environment by causing the bacteria to burst open (lyse)
Rationale of the experiment
By using the phage to lyse the bacteria, they could potentially deliver therapeutic proteins (like GFP) produced inside bacteria, to the gut more effectively
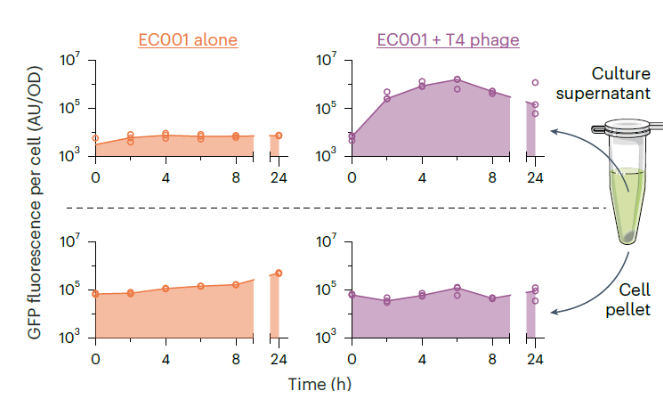
Experimental: With phage
significantly increases GFP fluorescence in the supernatant over time, indicating effective release of GFP due to phage-induced lysis, while GFP fluorescence in the cell pellet decreases as the bacteria lyse
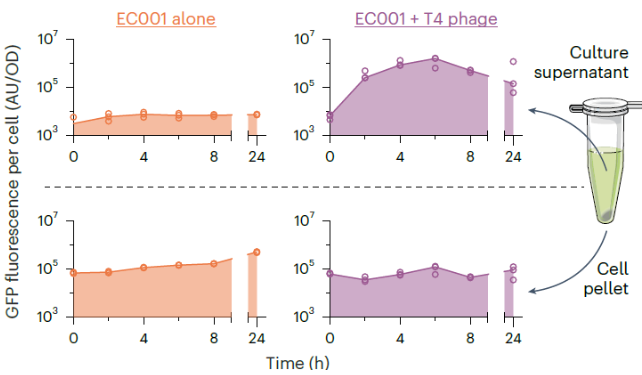
Control: Without phage
GFP fluorescence remains high in the cell pellet and low in the supernatant, indicating that GFP is contained within the bacterial cells without phage-induced lysis
AU/OD is arbitrary units/optical density. Why did they divide AU by OD?
the data reflect the amount of protein released per cell, rather than being influenced by the total number of cells
Transcription from phage promoters occurs in waves, first “early” promoters are turned on, then “middle”, then “late” promoters.
What class/function of genes are commonly transcribed at each stage?
Early promoters transcribe genes for DNA replication and host modification, middle promoters for DNA replication and recombination, and late promoters for structural proteins and lysis enzymes
How did they determine which promoter should encode the inserted foreign gene in the phage?
They tried promoters in the early, middle, and late stage of phage reproduction and chose the one with the maximum gene expression but lowest phage propagation impact
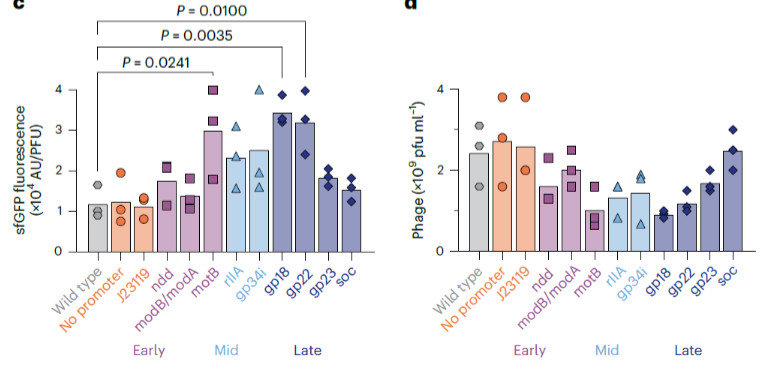
Given the chosen parameters (maximal sfGFP levels and minimal impact on phage titer) which promoter would you use and why?
gp22 is an early promoter that enables high expression of sfGFP while minimizing the effect on phage propagation.
Conclusion of experiment 1
Engineered T4 phage can orchestrate the production and release of heterologous proteins in the mammalian gut during the phage infection process, leveraging its co-existence with its bacterial host for a sustained in situ effect
Chimeric infective particles expand species boundaries in phage inducible chromosomal island mobilization
capsid-forming PICIs (cf-PICIs) only require what from helper phages
They only need tail proteins because they encode their own capsid
What is the main function of phage tail proteins
mediate the attachment and infection of host cells, facilitating the transfer of genetic material during the infection process.
SaPI vs cf-PICIs
Which are only found in S. aureus and have very narrow host genera?
SaPIs
SaPI vs cf-PICIs
Which are highly abundant and have a diverse group of hosts across species and genera?
cf-PICIs
cf-PICIs are highly abundant, and
often identical in sequence, across different
species and genera. What does this mean
suggest that these elements are highly efficient at horizontal gene transfer, allowing them to spread virulence and antibiotic resistance genes widely and rapidly
Tail-less cf-PICIs were isolated from the supernatant of induced Kp DSM30104. In addition, supernatants were isolated from induced E. coli isolates containing capsid mutant prophage (either HK106 or HK022, which have different tails). Why capsid mutants?
To make sure the prophage can only donate a tail
What was the purpose of experiment 2
To see if tail from one species specific phage will impact the ability of a different prophage to infect its host.
What was the purpose of experiment 3:
Natural inter-species cf-PICI transfer
transfer of the cf-PICI can occur naturally in mixed bacterial populations, but it requires a compatible helper phage (like HK022) to provide the necessary tails for the cf-PICI capsids
You hypothesize that the gene encoding a therapeutic protein could be encoded into a lytic phage so that it is co-expressed with phage genes during the infection of a patient’s resident gut bacterium, and that phage–bacterial co-existence would lead to the sustained production of your protein. Which of the options below would be best if the goal is to treat your patient?
High concentrations of your protein and of phage particles are released from each lysed bacterial cell.
Positive-sense (+) viral RNA
is like mRNA and can be immediately translated by host ribosomes
Negative-sense (-) viral RNA
is complementary to mRNA and must be converted
to (+) RNA before it can be translated
Baltimore groups 1 and 2 DNA viruses require
RNA polymerase to transcribe genome and a DNA polymerase to replicate genome
Baltimore group 7 DNA viruses require
RNA polymerase to transcribe genome AND a virus encoded RT to replicate genome
Large DNA viruses vs small DNA viruses:
Which ones DO NOT encode their own polymerases?
Small DNA viruses do not encode their own polymerases.
Baltimore group 6 and 7 (DNA) viruses require
a virus-encoded reverse transcriptase (RT) to replicate their genomes,
Baltimore groups 3, 4, and 5 RNA viruses require:
Virus-encoded RdRp (an RNA-dependent RNA polymerase) to both transcribe and replicate their genome
When is RdRp required following infection by a virus with a (+) ssRNA genome?
RdRp is not packaged within the virion. Instead, the viral genome is directly translated by the host cell's machinery to produce RdRp after infection
When is RdRp required following infection by a virus with a (-) ssRNA genome?
RdRp must be packaged within the virion because the host cell cannot directly translate the (-) ssRNA genome. The RdRp transcribes the (-) ssRNA into (+) ssRNA, which can then be translated into viral proteins and serve as a template for genome replication
Quasispecies
Virus populations are not identical, even within a single host, rather they exist as dynamic distributions of nonidentical but related replicons
How do latent herpesviruses persist in the host cell nucleus without being detected?
combination of histone modifications and regulatory RNAs inhibit viral transcription to limit expression of viral proteins
Marek's disease virus (MDV)
Herpes virus that infects chickens
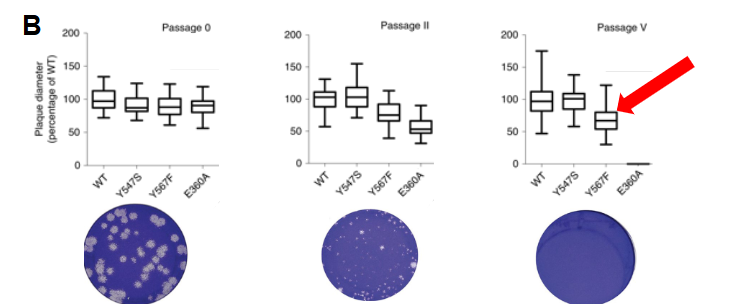
How would you interpret the cell culture data below?
Disrupting MDV DNA pol exonuclease activity reduces plaque size so MDV shows dependency on DNAP proofreading for effective infection.
provirus
dsDNA moves into the nucleus and inserts into the host genome
Quasispecies in regard to HIV
It takes 5-10 years to develop AIDS, during which time
HIV evolves into patient-specific quasispecies
Early stage (R5; Macrophage trophic) HIV virions
can infect dendritic cells and macrophage that display
CD4 (with CCr5 as co-receptor), and T cells that
display the same
Evolved late stage (X4; T-cell trophic) HIV
initiate infection using CD4 with CXCr4 as a co-receptor (only T cells)
What happens when viruses replicate above
their error threshold?
(1) The population contains too many mutations
and results in dead viruses.
(2) The population does not contain mutations
and results in live viruses.
(3) The population contains too many mutations
and produces more pathogenic viruses.
(4) The population cannot undergo reassortment
and produces less pathogenic viruses.
(5) The population does not contain mutations
and produces more pathogenic viruses
The population contains too many mutations
and results in dead viruses
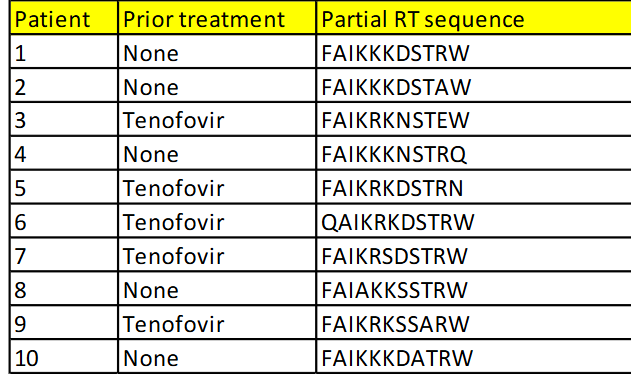
The table shows HIV genotyping data
from ten patients. HIV in the five
patients previously treated with
tenofovir have become resistant to this
base analogue inhibitor of RT. HIV in
the five new patients (untreated) are
sensitive to tenofovir. What is the RT
amino acid change that is associated
with the development of resistance?
Which of the following describes an activity provided by reverse transcriptase?
(A) It has protein-dependent RNA polymerase activity
(B) It has DNA-dependent RNA polymerase activity
(C) It has RNase H activity that hydrolyzes the RNA strand in an RNA/DNA hybrid
(D) It converts select proteins into their specific mRNA sequence
(E) It has RNA-dependent RNA polymerase activity
It has RNase H activity that hydrolyzes the RNA strand in an RNA/DNA hybrid
Which 3 functions does RT have
RNA-dep DNA pol
DNA-dep DNA pol
RNAse H activity
Your boss gives you $10 million to make Kirkland insulin. Q1: How would you proceed?
(1) Party like it’s 1922 (i.e., extract bovine insulin from pancreases)
(2) Party like it’s 1995 (i.e., clone Babe [the pig from the movie] and isolate his insulin)
(3) Party like it’s 2025 (i.e., just stand in the corner looking at your phone)
(4) Party like it’s 1978 (i.e., clone the insulin ORF into E. coli and overexpress/purify)
(5) Make the hotdogs from 100% bovine pancreases (....probably already are anyway)
Party like it’s 1978 (i.e., clone the insulin ORF into E. coli and overexpress/purify)
Q2: Which of the below is NOT something you need to worry about with regards to using E. coli
to express insulin?
(1) improper protein folding in E. coli
(2) loss of the expression plasmid from the E. coli cell
(3) intracellular accumulation of expressed proteins as inclusion bodies in E .coli
(4) alternative codon decoding rules between humans and E. coli
(5) lack of appropriate post-translational modifications in E. coli
alternative codon decoding rules between humans and E. coli
Plasmid vector component: MCS
sequence containing multiple unique restriction enzyme sites for easy foreign DNA insertion
Plasmid vector component: T7 promoter
drives high-level expression of a gene of interest in systems using T7 RNA polymerase, commonly used in protein production
Plasmid vector component: HIS-Tag
histidine residues added to proteins for purification purposes using metal affinity chromatography.
Q5: How could we test this is the
seq of the correctly spliced gene?
use primer 5’-ctggttcaagggcttta-3’
in conjunction with reverse
transcriptase to make a cDNA copy of
the mRNA, clone it, and then
sequence it
Q7: What do you need to
check before committing
to using NcoI and XhoI?
(1) that these enzymes do
not inhibit reverse
transcriptase activity
(2) that NcoI only cuts
phosphorylated DNA
(3) that a XhoI site is
present within the
antibiotic resistance gene
of the plasmid chosen for
overexpression
(4) that these enzymes do
not cut internal of your
gene of interest
(5) that these enzymes
function on DNA and not
RNA
That these enzymes do
not cut internal of your
gene of interest
A potential issue with producing a human protein in E. coli
involves the codons used
Human genes often use codons that are less frequently used or recognized by E. coli, leading to inefficient translation and lower protein expression
we’ve mentioned silent (aka. synonymous)
mutations a few times in this course. Q9: Do silent mutations
have any phenotype?
Maybe, while they change a gene codon to one that still
encodes the same amino acid, not all codons are equal and
hence there may be a change in phenotype
What could you do to boost insulin production in E. coli?
synthesize a codon-optimized version of the insulin gene
You add isopropyl β-D-1-thiogalactopyranoside (IPTG) to 0.5 mM. Q12: Group E: What is this and why do we add it?
molecular analog of allolactose used to induce gene expression in strains with the DE3 prophage, leading to the activation of T7 RNA polymerase and subsequent transcription of the insulin gene
Why do we run samples from the different steps (U, I, P, S, Ni-NTA, SEC) on the gel rather than just the SEC sample since that is the important one?
ensures each stage of the purification process is working correctly and helps identify any issues
When attempting to overexpress and purify a protein in E. coli it is common to use an IPTG-based expression system, where the gene of interest is only transcribed following addition of IPTG to the culture. Why is the gene of interest not significantly expressed until IPTG addition?
(A) IPTG inhibits the phage lysin protein present within pLysS; this prevents cell lysis and therefore enables the expression of the protein of interest
(B) IPTG binds to and inhibits the activity of LacI which results in the transcription of the T7 polymerase gene, which in turn results in the transcription of the gene of interest which is located downstream of a T7 promoter
(C) IPTG enables the E. coli to utilize sucrose as an energy source; the greater energy input results in enhanced protein production
(D) IPTG binds to lacZ which promotes the ability of the encoded protein, LacZ, to bind to the LacO region of the gene of interest’s promoter, enhancing transcription
(E) IPTG promotes the activity of the T4 DNA polymerase; resulting in enhanced transcription and translation of the gene of interest
IPTG binds to and inhibits the activity of LacI which results in the transcription of the T7 polymerase gene, which in turn results in the transcription of the gene of interest which is located downstream of a T7 promoter
Protozoa: Amoeba example
Brain eating amoeba called N. fowleri
Does N. fowleri have rRNA operons
No, each N. fowleri cell has ~4,000 copies of a 14 kb circular plasmid that harbor rRNA genes.
Protozoa: Flagellates example
T. brucei which causes African sleeping sickness and transmitted by tsetse fly
VSG switching by T. brucei
variant surface glycoprotein expression to create antigenetic variation for immune evasion
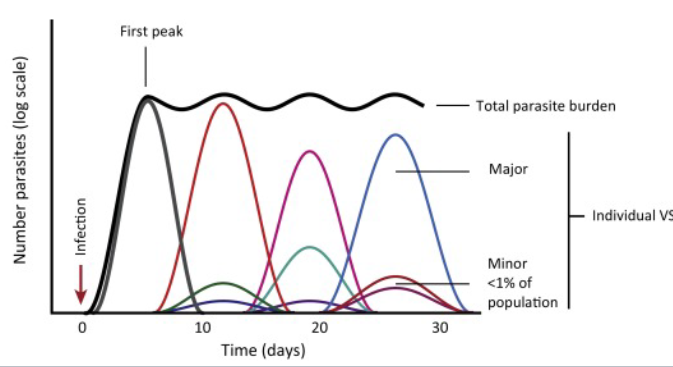
VSG switching by T. brucei pattern
Variants arise with degree of regulation because antibodies act to remove early variants, but by then later variants are already emerging
VSG switching mechanism
altering which 15 BES within the genome is active
recombination or gene conversion-mediated feeding of new
VSG sequence
Variant surface glycoprotein (VSG) switching is the major
driver of antigenic variation in Trypanosoma brucei. One
mechanism of VSG switching is what?
(1) inversion of the bloodstream expression site (BES) promoter
such that it drives transcription of a VSG gene located upstream
in the opposite orientation
(2) the mutation of RNA polymerase such that it recognizes a
different gene promoter sequence, which switches which VSG
gene is transcribed
(3) inducing transcription of a VSG gene located within one of
the densely-packed tandem arrays of VSG genes located within
one of the mini-chromosomes
(4) differential gene expression in response to levels of
extracellular iron
(5) the gene conversion-mediated feeding of new VSG sequence
information into the expressed bloodstream expression site (BES)
the gene conversion-mediated feeding of new VSG sequence
information into the expressed bloodstream expression site (BES)
Plasmodium life cycle: Schizogany
asexual reproduction by
multiple fission, producing merozoites.
The merozoites are what generate the
clinical symptoms of malaria
The first Plasmodium chloroquine-resistant isolate was recovered in 1957. In 2025, how could you determine the molecular basis of resistance?
Metagenomic analysis to compare chloroquine-resistant and -sensitive isolates
What happened when comparing chloroquine-resistant and -sensitive isolates of P. falciparum?
identified a gene on chromosome 7 as the major resistance determinant
Hookworm genomics
shed some light on the remarkable ability of these organisms to
suppress the production of pro-inflammatory cytokines
Female schistosoma
heterogametic sex with ZW chromosome
Male schistosoma
Homogametic with ZZ pair
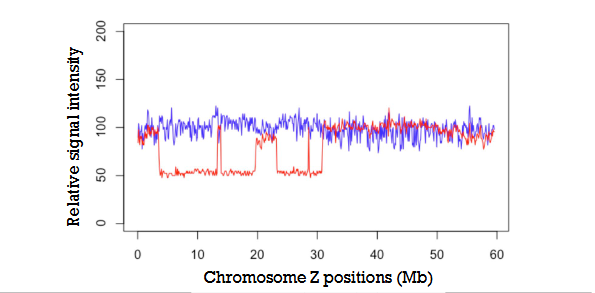
Analysis of male and female sex chromosomes in Schistosoma
suggests that there are distinct regions on the Z chromosome that differ between males and females, which could be indicative of sex-specific genetic variations or differences in chromosome structure and gene content
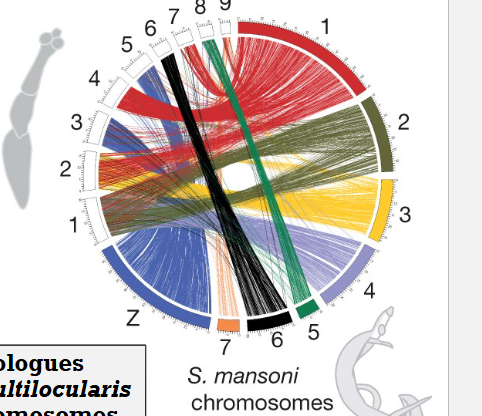
Tapeworm genome vs Flatworm distant relative
Although gene order has been lost, ancient chromosomal synteny is preserved among these parasitic flatworms
Genome data has been generated for several
species of hookworm. One insight to come
out of this data is what?
(1) insight into the mechanisms by which these
parasites can suppress production of pro-
inflammatory molecules
(2) observation that these parasites do not
express proteins on their surface, and hence no
vaccine can be developed
(3) observation that the rRNA operons of these
parasites are located on plasmids not as part of
one or more chromosomes
(4) that different hookworm species are
unrelated to each other, sharing as much
similarity to C. elegans as they do with each other
(5) that sequencing the genomes of these
multicellular eukaryotes is easy
insight into the mechanisms by which these
parasites can suppress production of pro-
inflammatory molecules
S. cerevisiae mating pheromones
a-factor
α-factor
S. cerevisiae: a cells
respond to α-factor by growing a projection towards
the source of α-factor
Gene organization in the MAT, HML, and HMR loci on S. cerevisiae
chromosome III
MAT locus contains expressed mating allele, but HML and HMR contain silenced copies of a and α so that mating type switching can occur
MAT locus
region on chromosome III responsible for the expressed mating allele in S. cerevisiae.
HML locus
region on chromosome III containing silenced copies of silenced copy of MATα in S. cerevisiae.
HMR locus
region on chromosome III containing silenced copy of MATa in S. cerevisiae.
Which of the S. cerevisiae strain pairs listed below will result
in shmoo formation toward each other?
(1) two diploid cells (one which is a/α, and the other which is α/a)
(2) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes a-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes α-
factor
(3) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a a/α diploid cell
(4) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes a-
factor
(5) a haploid “a” cell that produces α-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces a-factor and expresses a receptor that recognizes a-
factor
haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes a-
factor
Gal4 bait and prey technique is used to determine
If a protein interacts with 2 test proteins
Gal4 bait and prey overview
create a bait construct with the Gal4 binding domain fused to your protein and two prey constructs with the Gal4 activation domain fused to each test protein; co-transform these into S. cerevisiae with a reporter gene, and if the bait and prey proteins interact, the reporter gene is transcribed
In the gal4 technique, what do you fuse the “favorite protein” to
The gal4 binding domain
In the gal4 technique, what do you fuse the non-favorite protein to
the Gal4 activation domain
In the gal4 technique, if the bait and prey interact, then what happens
the reporter gene is transcribed.
In the gal4 technique, if the bait and prey don’t interact, then what happens
the reporter gene is not transcribed.
How come most organisms have only 45unique tRNAs
the 3rd base pairs by wobble rules so that fewer codons are needed
Wobble rules
Inosine in the anticodon can pair with uracil (U), cytosine (C), or adenine (A) in the codon, allowing for flexible base pairing
G-U
Decoding CUN in S. cerevisiae
only uses two tRNAs to translate the four CUN codons
(each of which translates two codons)
Candida CUN decoding overall
Candida species translate CUG codons as serine instead of leucine
Candida CUN decoding mechanism
dedicated tRNACAGSer for CUG codons and a single tRNA IAGLeu for CUA, CUC, and CUU codon
Mycorrhiza
A symbiotic association between fungi and plant roots that enhances nutrient exchange.
endomycorrhizas
occur in ~80% of all plants. The fungal hyphae penetrate the cell walls of plant root cells and invaginate the cell membrane to create a greater contact surface area, promoting nutrient transfer
ectomycorrhizas
occur in ~10% of all plants, particularly “woody” plants (e.g., trees). The hyphae of these fungi do not penetrate individual cells within the plant root, rather they make a sheath around roots
Candida species use only two different tRNAs to
decode all four CUN codons (where N is any
nucleotide). How is this possible?
(1) the third base of an anticodon can pair by the
wobble rules (i.e., G-U, I-U, I-A, I-C)
(2) the third base of a codon shows no specificity of
binding
(3) the second base of an anticodon can pair
simultaneously with the second and third bases of a
codon
(4) the third base of an anticodon can pair by the
womble rules (i.e., G-A, I-U, I-A, I-G)
(5) the third base of an anticodon can pair by the
wobble rules (i.e., G-C, I-U, I-A, I-C)
the third base of an anticodon can pair by the
wobble rules (i.e., G-U, I-U, I-A, I-C)
Do all beer yeast make the same metabolites
NO, different strains of beer yeast produce varying metabolites, affecting flavor, aroma, and alcohol content.
Why do yeast strains differ in their alcohol tolerance
allows yeast to survive and continue fermenting in environments with high alcohol concentrations, which is crucial for producing alcoholic beverages and biofuels
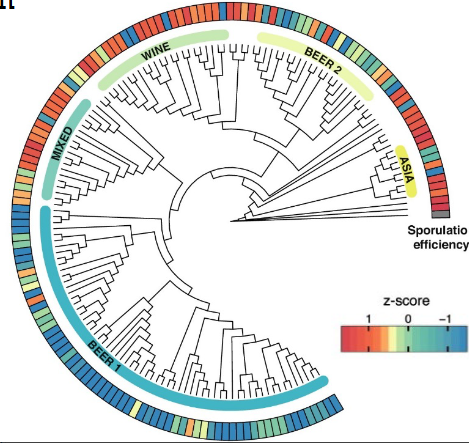
Diversity of sporulation efficiency in
industrial S. cerevisiae strains
Beer yeasts have lost their ability to effectively sporulate because they are only exposed to nutrient rich environments
Candida albicans and close relatives harbor a unique barrier to horizontal gene transfer that is not seen in other fungal species. What is this barrier?
(A) they have no cell surface glycoproteins and hence lack receptors required for viral infection
(B) they only grow as spores and hence, due to the thick spore coat, are protected against DNA entry
(C) they harbor an unusually large number of R-M complexes within the periplasm
(D) they harbor uracil, not thymine, in their DNA
(E) they use a non-standard genetic code
E. They translate CUG codons as serine (while most other organisms on the planet translate as leucine). Thus, if genes are introduced into C. albicans (let’s say from S. cerevisiae), and these genes have CUG codons in them, these will be translated as serine which may lead to unfunctional proteins
Archaea are most similar to
Eukaryotes with a single membrane and thick cell wall
Archaea are resistant to a wide variety of antibiotics that are primarily produced by
Gram positive bacteria
Why are archaea resistant to gram positive produce antibiotics
these antibiotics primarily act on factors that distinguish
archaea from bacteria
Do archaea have more or less introns than bacteria
More because they are more complex