Human Genetics Exam 2 Study Guide
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
49 Terms
What is genetic linkage?
Genetic linkage occurs when genes are located on the same chromosome. Alleles on the same chromosome tend to be inherited together unless crossing-over occurs between homologous chromosomes during meiosis.
What are recombinant offspring?
Recombinant offspring are those that have an arrangement of alleles on the chromosome that was not present in either parent.
Genes X and Y are located 2 m.u. apart on a chromosome and genes Y and Z are located 8 m.u. apart. Which genes will have the highest recombination frequency- X and Y or Y and Z?
Genes Y and Z because they are located further apart. Recombination frequency is equal to map distance, so genes X and Y will have an expected recombination frequency of 2% and Y and Z will have an expected recombination frequency of 8%.
We have 2 genes, gene A and gene B. If a parent heterozygous for both genes (AaBb) is crossed to a parent homozygous recessive for both genes (aabb), how will the expected genotype ratios of offspring differ if the genes are assorting independently versus if the genes are linked?
If the genes are assorting independently, we would expect: 25% of offspring to be AaBb, 25% of offspring to be aabb, 25% of offspring to be Aabb and 25% of offspring to be aaBb.
If the genes are linked, two genotypes will be present more than 50% of the time, and the other two genotypes will be present less than 50% of the time. We can’t say which genotypes will be more than 50% and which genotypes will be less than 50% without knowing the arrangement of alleles on the chromosomes of the heterozygous parent (i.e., A___B a___b or A___b a___B).
Two genes are located on the same chromosome and are known to be 12 m.u. apart. An AABB individual is crossed to an aabb individual to produce AaBb F1 offspring. The F1 offspring were then crossed to an aabb individual.
If 1000 F2 offspring were produced from this cross, what are the predicted numbers of F2 offspring with each of the 4 genotypes: AaBb, Aabb, aaBb and aabb?
Parents are A___B A___B and a___b a___b.Therefore, the F1 offspring is A___B a___b.The F1 is crossed to a tester: i.e., A___B a___b x a___b a___b The non-recombinant F2 offspring are:A___B a___b and a___b a___b
(AaBb) (aabb)The genes are 12 m.u. apart so there will be 100-12 = 88% non-recombinant F2 offspring. 88% of 1000 is 880. Therefore, 440 offspring will be A__B a__b and 440 will be a__b a__b The recombinant F2 offspring will be:A___b a___b and a___B a___b
(Aabb) (aaBb)
The genes are 12 m.u. apart so 12% of offspring will be recombinant;12% of 1000 is 120. Therefore, 60 will be A___b a___b and 60 will be a___B a___b
Two genes are located on the same chromosome and are known to be 12 m.u. apart. An AABB individual is crossed to an aabb individual to produce AaBb F1 offspring. The F1 offspring were then crossed to an aabb individual.
What would be the predicted numbers of F2 offspring with these 4 genotypes if the parental generation had been AAbb and aaBB instead of AABB and aabb?
The parents are A___b A___b and a___B a___B
Therefore, the F1 offspring would is A___b a___BThe F1 is crossed to a tester: i.e., A___b a___B x a___b a___b The non-recombinant F2 offspring areA___b a___b and a___B a___b
(Aabb) (aaBb)The genes are 12 m.u. apart, so there will be 100-12 = 88% non-recombinant offspring 88% of 1000 = 880, so 440 will be A__b a__b and 440 will be a__B a__bThe recombinant F2 offspring are:A___B a___b and a___b a___b
(AaBb) (aabb)The genes are 12 m.u. apart so there will be 12% recombinant offspring. 12% of 1000 is 120, so 60 will be A__B a__b and 60 will be a__b a__b
Huntington disease: H = dominant; h = normal
Molecular marker (MM) alleles:
MMa1 = molecular marker allele 1; MMa2 = allele 2; MMa3= allele 3
In 50 families analyzed, one parent is H___MM a2 and the other parent is h___MM a1 h ___MM a3 h___MM a1
In 50 children, 20 have the Huntington allele (H) and molecular markers MM a1 and MM a2; 20 have 2 normal alleles (h) and molecular markers MM a1 and MM a3; 5 have the Huntington allele (H) and molecular markers MM a1 and MM a3 and 5 have 2 normal alleles (h) and molecular markers MM a1 and MM a2.
What is the recombination frequency?
Recombination frequency (RF) = 10/50 = 0.2 = 20%
Huntington disease: H = dominant; h = normal
Molecular marker (MM) alleles:
MMa1 = molecular marker allele 1; MMa2 = allele 2; MMa3= allele 3
In 50 families analyzed, one parent is H___MM a2 and the other parent is h___MM a1 h ___MM a3 h___MM a1
In 50 children, 20 have the Huntington allele (H) and molecular markers MM a1 and MM a2; 20 have 2 normal alleles (h) and molecular markers MM a1 and MM a3; 5 have the Huntington allele (H) and molecular markers MM a1 and MM a3 and 5 have 2 normal alleles (h) and molecular markers MM a1 and MM a2.
How many map units apart is the Huntington allele from the molecular marker?
Therefore, the Huntington allele is 20 m.u. from the molecular marker.
For this question, use the map distance you calculated in question 6b. Beth has a father who has developed Huntington disease and a mother that definitely does not have Huntington disease. Beth has molecular markers MMa2 and MMa3. Given the genotypes of her mother and father below, what is the probability that Beth will develop Huntington disease?
Beth’s father: H___MMa2 h___MMa1
Beth’s mother: h___MMa1 h___MMa3
MMa2 is linked to the Huntington allele. Beth must have received the MMa2 allele from her father. As her mother is unaffected, we need only consider whether or not Beth inherited the Huntington allele from her father.
From the father, Beth could have received H___MMa2 (non-recombinant) or h___MMa2 (recombinant). (We know that she did not receive H____MMa1 or h___MMa1).
The genes are 20 m.u. apart, so the probability that Beth got the recombinant h___MMa2 allele is 20% (we know she did not get H___MMa1 so that probability is zero). Therefore, the probability that Beth got the non-recombinant H___MMa2 allele combination is 80% (we know she did not get h___MMa1 so that probability is zero).
The gene for nail patella syndrome (N), a rare autosomal dominant trait, is 10 m.u. apart from the I gene that determines the ABO blood group.
Greg and Susan each have nail patella syndrome. Greg has blood type A and Susan has blood type B.
(Greg’s mother has nail patella syndrome and blood type A) (Greg’s father has normal nails and knees and blood type O) (Susan’s mother has nail patella syndrome and blood type O) (Susan’s father has normal nails and knees and blood type B)
What is the probability that their child will have normal knees and nails and blood type B?
Gregn___i (from Father) and N___IA from mother SusanN___i (from Mother) and n___IB from father
ChildMust get n___i from Greg (non-recombinant)- 0.45 chance of getting this non-recombinant chromosomeMust get n___IB from Susan (non-recombinant)- 0.45 chance of getting this non-recombinant chromosome
Therefore, 0.45 x 0.45 = 0.20 or 20%
A molecular marker is located 2 map units away from the X-linked locus responsible for red- green color blindness. Three alleles of this molecular marker are known: "3 repeats," "5 repeats," and "7 repeats." A woman with normal color vision, but whose father is color blind, has a partner with normal vision, and they have a son.
Given the molecular marker genotypes of the various family members shown above, what is the probability that the couple's baby son is color blind?
The genes are X-linked, so recombination can only occur in the mother.
The woman’s father is colorblind and has 5 repeats, so the colorblind allele is linked to the 5 repeat molecular marker (a_____5), which the woman received from her father.
The child also has 5 repeats. The only way the son is not colorblind is if there was crossing-over between the woman’s X-chromosomes. The 2 genes are 2 m.u. away, so the chance of a cross- over event is 2%. Therefore, the chance the son is colorblind is 100 – 2 = 98%

What is meant by extranuclear DNA, and where is extranuclear DNA located?
Extranuclear DNA refers to DNA located outside the nucleus. Extranuclear DNA is located in mitochondria and also in chloroplasts in plants.
In animals, what type of inheritance does mitochondrial DNA (mtDNA) show, and why does it show that inheritance pattern?
Maternal inheritance because it is the egg cell that provides all of the mitochondria to a zygote. The sperm cell contributes negligible mitochondria to the zygote.
What is meant when we say that mitochondria are semiautonomous?
It means that mitochondria contain some of their own DNA, that is, DNA that encodes some of the proteins and RNAs that function in mitochondria. However, a mitochondrion does not encode all of its proteins; most of the proteins that function in the mitochondrion are encoded by the nucleus.
Amanda's mother was mildly affected with the mitochondrial disorder NARP (neurologic muscle weakness, ataxia and retinitis pigmentosum). Amanda is phenotypically normal, so she decides to have children. Her first child is mildly affected with NARP, and her second child is severely affected with NARP. How can two siblings from the same parents display such varying expressivity?
Due to heteroplasmy- the ratio of normal to mutant mitochondria in a cell. The second child may have developed from a zygote that inherited a higher ratio of affected to normal mitochondria from the mother.
Tom is mildly affected with the mitochondrial disorder MERRF (myoclonic epilepsy and ragged red fiber disease), which he knows is due to a mutation in his mtDNA. His partner is phenotypically normal and does not have a family history of mitochondrial disease. Do the couple need to be concerned about having children affected with the MERRF?
The couple should not be concerned about having a child with MERRF (that is, no more concerned than the average couple) because the child will inherit normal mtDNA from the mother.
Why can mitochondrial disorders be cause by mutations in mitochondrial DNA OR nuclear DNA?
Mitochondrial DNA encodes products (polypeptides and functional RNAs) that function in the mitochondria. Therefore, a mutation in mtDNA will affect those products. Additionally, the majority of products (polypeptides and functional RNAs) that function in the mitochondria are encoded my DNA in the nucleus (the products are transported to the mitochondria following translation); therefore, a mutation in a nuclear gene that encodes a product that functions in the mitochondria will also affect the function of the mitochondria.
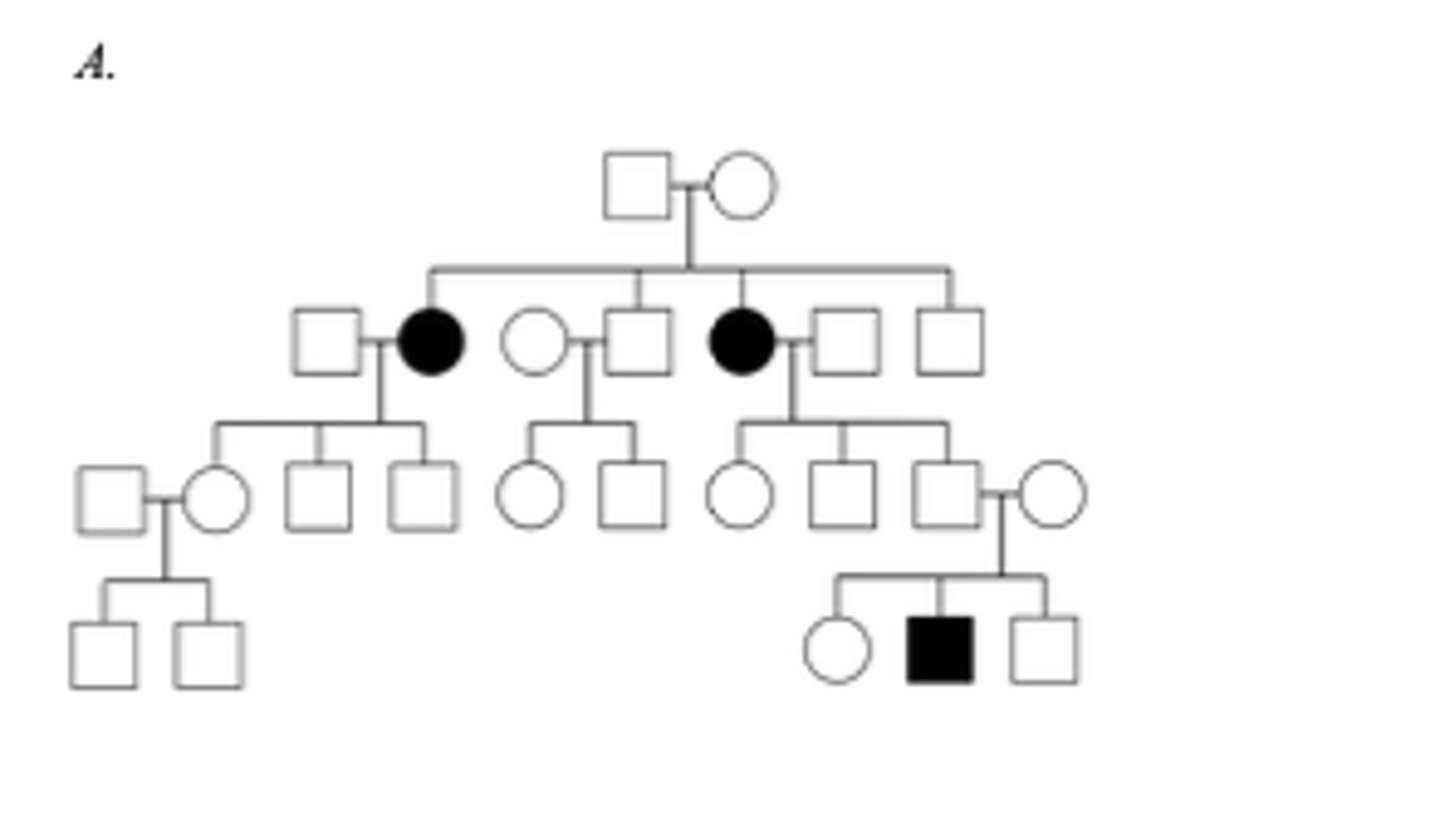
Below are three family pedigrees for different mitochondrial disorders. For each of the pedigrees A - C, is the mutation in mtDNA or nuclear DNA? A
The mutation is in nuclear DNA because it does not show maternal inheritance; its hows an autosomal recessive pattern of inheritance.
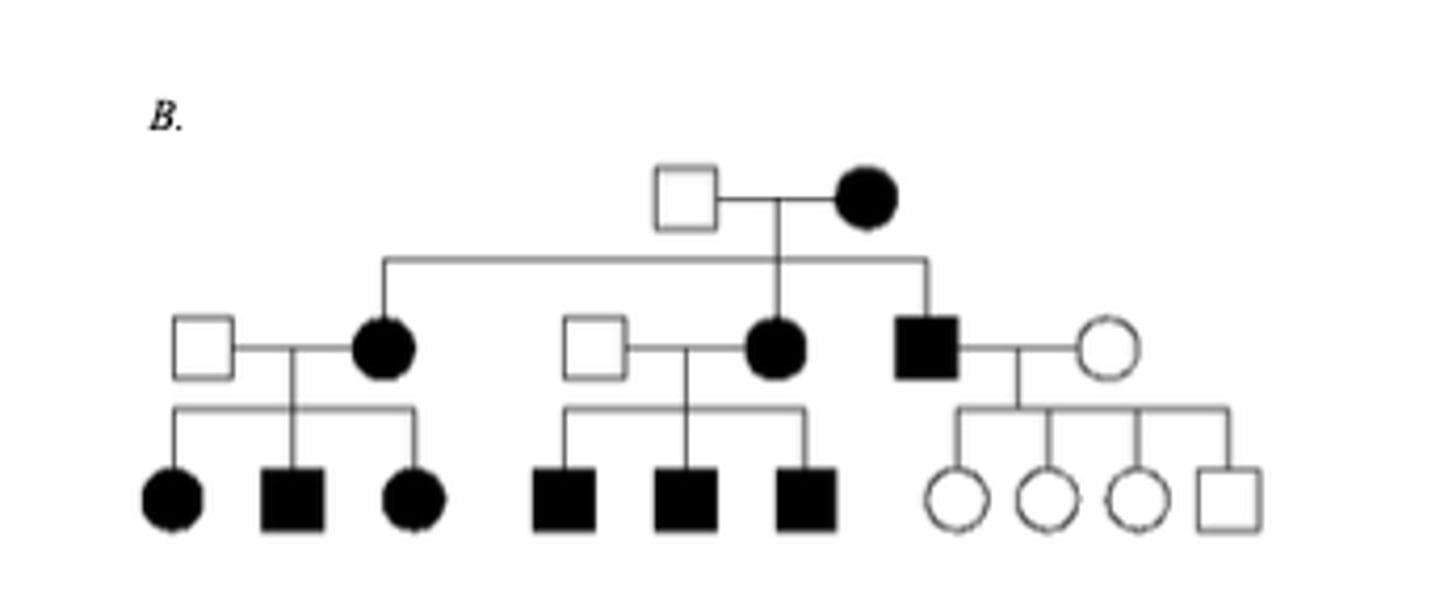
Below are three family pedigrees for different mitochondrial disorders. For each of the pedigrees A - C, is the mutation in mtDNA or nuclear DNA? B
The mutation is in mtDNA because it shows a maternal pattern of inheritance. Affected females have affected children and the affected male does not.
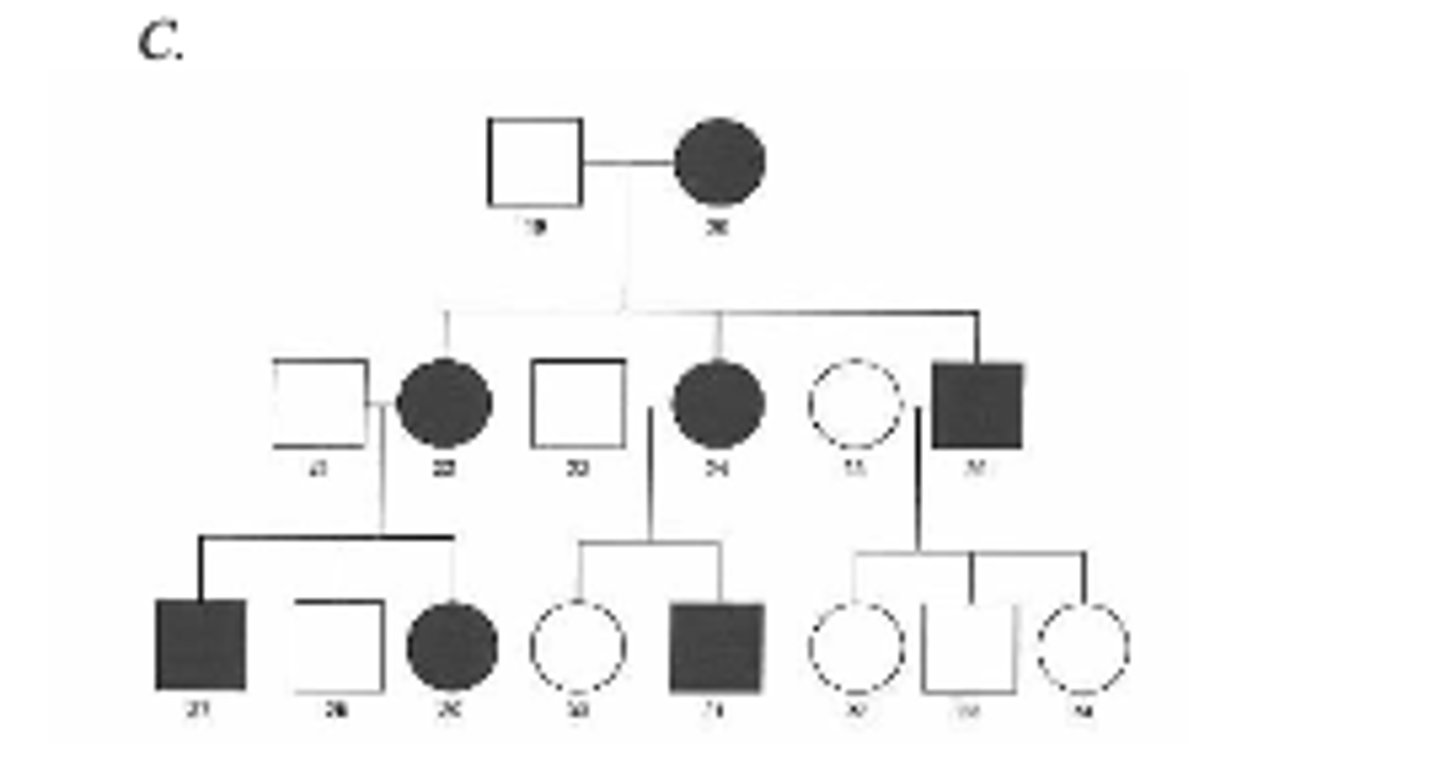
Below are three family pedigrees for different mitochondrial disorders. For each of the pedigrees A - C, is the mutation in mtDNA or nuclear DNA? C
Eitheranswerisacceptableforthisonewithoutknowingmoreinformation.
The mutation is likely in mtDNA because it seems to show a maternal pattern of inheritance with an affected female having affected offspring and an affected male having unaffected offspring. Individuals 28 and 30 may be phenotypically normal (although they have an affected mother) due to inheriting a sufficient ratio of normal versus mutant mitochondria to prevent disease symptoms. Alternatively, it could be autosomal dominant transmission with incomplete penetrance (although less likely).You will not get one a tricky as this on the exam- on the exam I would say to assume complete penetrance of any inherited mutations.
In phenotypes that show a maternal effect, what determines the phenotype?
The genotype of the mother determines the phenotype of offspring, because gene products in the mother affect the developing fetus.
What is epigenetic inheritance?
Gene expression is altered without any changes to DNA sequence.
In X inactivation, why do females inactivate one of their X chromosomes in all cells? Do females inactivate the maternal X or the paternal X? What is a Barr body?
Females inactivate one X in order to achieve dosage compensation, which means that males and females must have the same level of gene expression for genes located on the X chromosome because genes located on the X-chromosome are involved in the determination of traits other than sex. Because males only have 1 X, they only have 1 copy of genes located in the differential region of the X, so females must therefore inactivate one of their X chromosomes.
For X-linked recessive disorders, what is meant by the term 'manifesting heterozygote'?
Early in fetal development, the female randomly inactivates one X chromosome in all cells, so some cells will inactivate the maternal X and other cells will inactivate the paternal X.
The inactivated X chromosome becomes highly condensed (compacted), and a Barr body refers to the highly condensed inactivated X chromosome.
How can skewed X-inactivation affect the expressivity of X-linked conditions in females?
It means that a female who is heterozygous displays mild disease symptoms. On average, 50% of cells of a heterozygous female express the normal X and 50% of cells express the affected X, so it is not unusual for a heterozygous female to display mild symptoms. Due to skewed X-inactivation, a female may have <50% of cells expressing the normal X and >50% of cells expressing the affected X (in which case she will be more likely to display symptoms) or vice versa (in which case she will be less likely to display symptoms).
An individual has the genotype XXY. Is this individual male or female?
The individual is male. The Y chromosome determines whether an individual is male.
How many Barr bodies would the individual contain in somatic cells?
The male will have 1 Barr body in all cells as he will inactivate 1 of his X chromosomes.
Females with Turner syndrome are missing an X chromosome (i.e., they only have 45 total chromosomes in all cells). If females normally inactivate 1 X chromosome in all cells then why aren't Turner syndrome females phenotypically normal?
Females normally express genes in the pseudoautosomal region on both chromosomes. Genes located in the pseudoautosomal region of the X and Y show biallelic expression (both gene copies are expressed). Therefore, Turner syndrome females only have half the normal amount of gene expression for genes in the pseudoautosomal regions.
The alleles that determine black and orange fur in cats are located on the X chromosome. A tortoiseshell female cat comes home pregnant and later has kittens. 1 male kitten is orange, 2 male kittens are black, 2 female kittens are tortoiseshell and 1 female kitten is orange. What color was the male cat?
He must have been orange. This is the only way that an orange female with 2 orange alleles could have been produced.
What is meant by genomic imprinting? In maternal imprinting, which gene copy is imprinted, and which gene copy is expressed? In paternal imprinting, which gene copy is imprinted, and which gene copy is expressed?
It means that one gene copy in a gene pair is silenced. Genes that are maternally imprinted are imprinted if they come from the mother, and genes that are paternally imprinted are imprinted if they come from the father.
Beckwith-Wiedermann syndrome can result from a mutation in the CDKN1C gene, which is paternally imprinted. If a normal female (homozygous for the normal allele) and a male with Beckwith Wiedermann syndrome (heterozygous- 1 normal allele and 1 affected allele) have children:
What proportion of their offspring will be phenotypically affected with the syndrome?
None of their sons or daughters because the affected gene copy from the father will be imprinted.
Beckwith-Wiedermann syndrome can result from a mutation in the CDKN1C gene, which is paternally imprinted. If a normal female (homozygous for the normal allele) and a male with Beckwith Wiedermann syndrome (heterozygous- 1 normal allele and 1 affected allele) have children:
If their son gets the affected allele, what proportion of his children will be phenotypically affected with the syndrome (assuming his partner has normal alleles)?
None of their son’s children will be phenotypically affected because they will express the gene from their mother.
Beckwith-Wiedermann syndrome can result from a mutation in the CDKN1C gene, which is paternally imprinted. If a normal female (homozygous for the normal allele) and a male with Beckwith Wiedermann syndrome (heterozygous- 1 normal allele and 1 affected allele) have children:
If their daughter gets the affected allele, what proportion of her children will be affected with the syndrome?
If the daughter received the affected allele from her father then half of her children will be affected because half of her gametes will carry the affected allele, and the gene is expressed when it is inherited from a female. If the daughter did not receive the affected allele from her father then none of her children will be affected.
Beckwith-Wiedermann syndrome can result from a mutation in the CDKN1C gene, which is paternally imprinted. If a normal female (homozygous for the normal allele) and a male with Beckwith Wiedermann syndrome (heterozygous- 1 normal allele and 1 affected allele) have children:
How would the answers to parts a-c be different if it was an affected female (heterozygous) and a normal male (homozygous for the normal allele) in the parental generation?
Half of their sons and daughters would be affected because half of the mother’s gametes would be affected.
None of their son’s children would be phenotypically affected because they will express the gene from their mother.
If the daughter received the affected allele from her mother, then half of her children would be phenotypically affected because half of her gametes will have the affected allele.
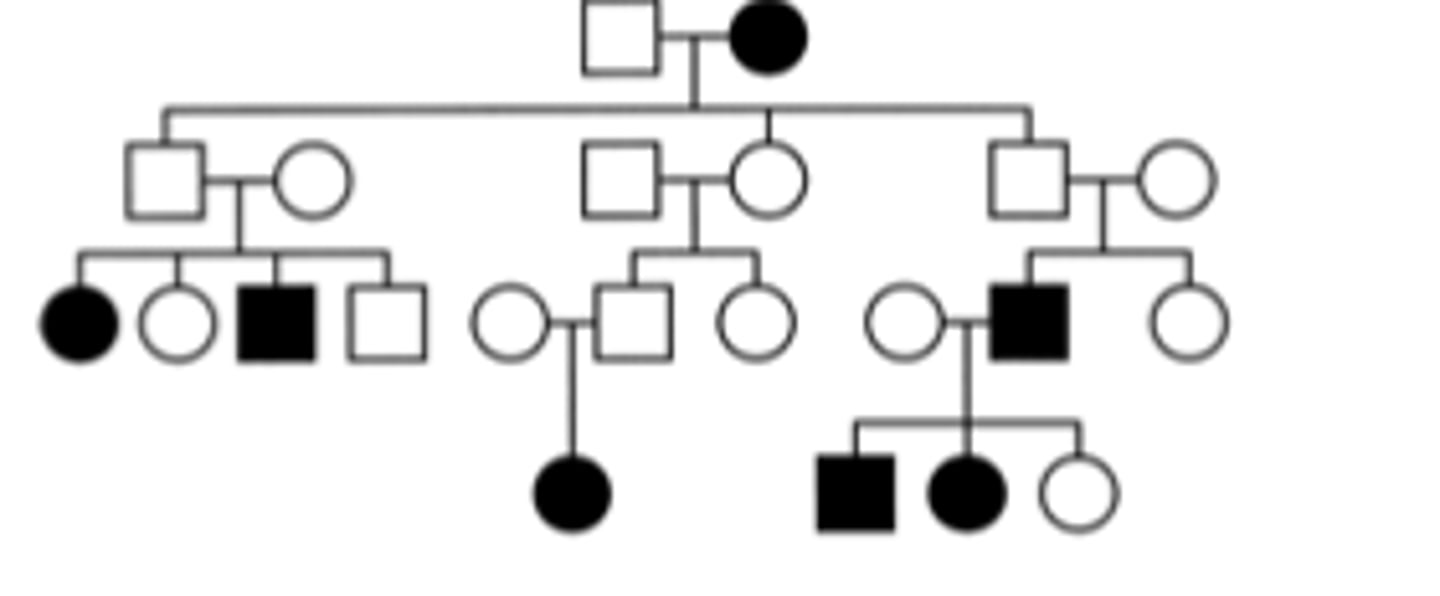
Does the pedigree below show inheritance of a disorder caused by a paternally imprinted gene or a maternally imprinted gene? Assume people marrying into the pedigree do not have an affected allele.
Maternal imprinting because the affected allele is expressed when it comes from males, but not when it comes from females. None of the children of the affected mother in generation 1 are affected but they are evidently carriers. Both of her sons have affected offspring, but her daughter does not. However, her daughter’s son is evidently a carrier because he has an affected daughter. Also, the affected male in generation 3 goes on to have affected offspring.
Approximately 75% of cases of Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are caused by a deletion of a small region on chromosome 15 that contains a cluster of imprinted genes. The gene that causes PWS is normally maternally imprinted, and the gen e that causes AS is usually paternally imprinted. If an individual has the chromosome 15 deletion on the paternal chromosome, which syndrome will resu lt? If an individual has the chromosome 15 deletion on the maternal chromosome, which syndrome will result?
PWS syndrome. The gene that causes PWS is usually maternally imprinted, so a paternal deletion means that there is no functional copy of the gene.
Because of the nature of the conditions, PWS and AS individuals do not have offspring of their own. However, hypothetically, say a male individual with AS (caused by a deletion of the AS/PWS genes on chromosome 15) did have a child with a normal female partner, what is the probability that the child would have PWS, and what is the probability that the child would have AS? Hypothetically, say a female individual with AS (caused by a deletion of the AS/PWS genes on chromosome 15) did have a child with a normal male partner, what is the probability that the child would have PWS, and what is the probability that the child would have AS?
Hypothetically, there would be a 50% chance that a male with AS would transmit the chromosome with the deletion, and, in that case, the resulting offspring would have PWS syndrome (the PWS gene normally shows paternal expression, so if that gene is not present on the paternal chromosome, then PWS results). There is a 0% chance an affected male would have a child with AS because the AS gene is expressed from the maternal chromosome.
Hypothetically, there would be a 50% chance that a female with AS would transmit the chromosome with the deletion, and, in that case, the resulting offspring would have AS syndrome (the AS gene normally shows maternal expression, so if that gene is not present on the maternal chromosome, then AS results). There is a 0% chance an affected female would have a child with PWS because the PWS gene is expressed from the paternal chromosome.
What are the 3 components of a nucleotide? Which type of sugar is found in DNA? Which type of sugar is found in RNA? Which 4 bases are found in DNA? Which 4 bases are found in RNA?
The components of a nucleotide are one or more phosphate groups, a pentose sugar (ribose in RNA nucleotides and deoxyribose in DNA nucleotides) and a base (thymine, adenine, cytosine or guanine in DNA nucleotides and uracil, adenine, cytosine or guanine in RNA nucleotides).
What is the difference between a base, a nucleoside, and a nucleotide?
A base is just a base, e.g., thymine, guanine, cytosine, adenine, uracil.A nucleoside is a base + sugar (e.g., thymidine, guanosine, cytidine, adenosine, and uridine) A nucleotide is a base + sugar + phosphate group(s) (i.e., thymidine triphosphate)
Which 2 components of nucleotides form the backbone of a DNA strand, and which component points towards the center to interact with the opposite strand?
The phosphate groups and sugar groups form the backbone of a DNA strand and the bases points towards the center where they hydrogen bonds with a bases on the opposite strand.
What are the complimentary base pairing rules in DNA and RNA?
In DNA, A pairs with T and G pairs with C In RNA, A pairs with U and G pairs with C
What type of bond links nucleotides together to form a single strand? What type of bond forms between bases on adjacent strands to hold together the two DNA strands in the double helix? How many bonds form between AT? How many bonds form between GC?
A phosphodiester bond links nucleotides together.Hydrogen bonds form between bases on opposite strands.Two hydrogen bonds form between an AT base pair, and 3 hydrogen bonds form between a GC base pair.
If a DNA strand has the sequence 5' - ACG TAG CCA GGT - 3', what is the sequence of the complimentary strand? (Remember DNA sequences are always conventionally written from 5' - 3'.
5’–ACCTGGCTACGT–3’ (this is the same thing as3’–TGCATCGGTCCA–5’)
If a DNA sample contains 20% C, what is the % of T?
30% (If there is 20% C then there is also 20% G. That leaves 30% T and 30% A).
In DNA and RNA nucleotides, what group is attached to the 5' carbon? What group is attached to the 3' carbon? In a DNA or RNA strand, what group is at the 5' end, and what group is at the 3' end?
A phosphate group is attached the 5’ carbon, so there is a free phosphate group at the 5’ end of a DNA/RNA strand. A hydroxyl (OH) group is attached to the 3’ carbon, so there is a free hydroxyl group at the 3’ end of a DNA/RNA strand.
What does it mean when we say the two strands in the DNA double helix run antiparallel?
One strand runs 5’ – 3’ and the other strand runs 3’ – 5’.
Give an explanation for why genome size does not correlate with the complexity of eukaryotic organisms.
he majority of eukaryotic genomes is composed of non-coding DNA (only ~1.5% of the human genome is composed of genes that encodes a functional product). Therefore, the eukaryotic genome size is determined more by the amount of non -coding DNA than by the number of functional genes that determine an organisms traits.
What is chromatin composed of? What is a nucleosome? What are the next two levels of DNA compaction after nucleosomes?
Chromatin is composed of DNA and histone proteins. Nucleosomes are the structural units of chromatin; they are composed of DNA wound around histone proteins. Nucleosomes are compacted into 30 nm fibers and 30 nm fibers are further compacted into radial loop domains.
What is the difference between euchromatin and heterochromatin?
Euchromatin is composed of less condensed DNA with radial loop domains being the highest level of compaction. Euchromatin tends to contain regions of DNA that have a high density of actively transcribed genes, so the chromatin is less condensed to enable the transcription machinery to access the DNA.Heterochromatin is composed of highly condensed DNA where radial loop domains are compacted even further. Heterochromatin tends to contain DNA that does not have a high density of actively transcribed genes, so therefore the transcription machinery does not need to be able to access the DNA.