Module 9: Cell Cycle Regulation
1/78
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
79 Terms
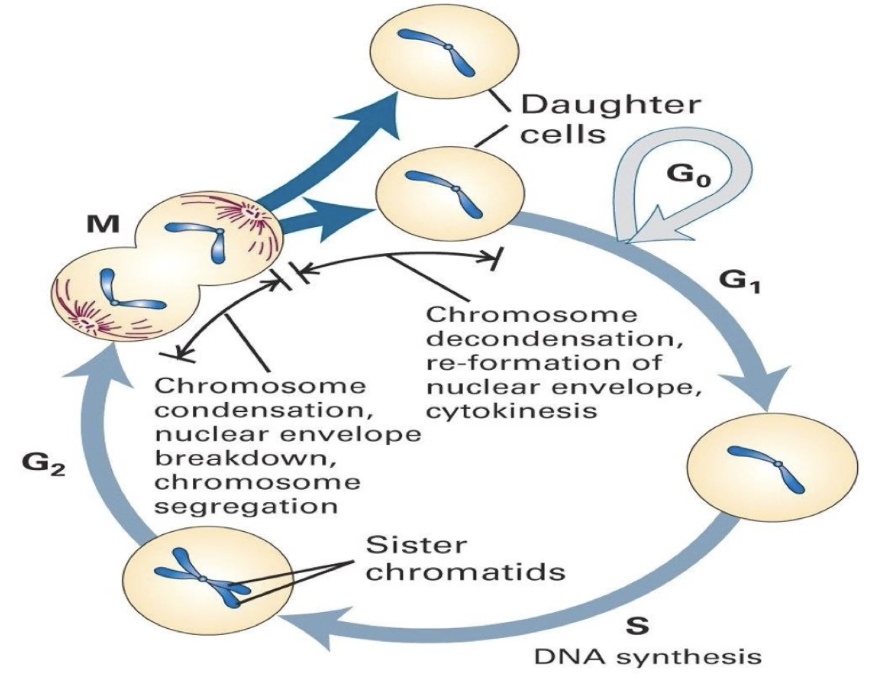
What are the main phases of the cell cycle?
G1: Cell grows, gene expression, protein synthesis
S: DNA replication → sister chromatids formed
G2: Prepares for mitosis
M: Mitosis → divides chromosomes into daughter cells
G0: Quiescent state (non-dividing)
Why is cell cycle regulation important?
Ensures DNA replication and division alternate properly
Prevents cell death or over-proliferation (e.g., cancer)
Most cells are in G0, not actively dividing
What can go wrong with poor regulation?
Cell death if replication/division fail
Over-proliferation → cancer
Tissue cannot be repaired if division stops
What are the possible fates of a stem cell entering the cell cycle?
Self-renewal → 2 identical stem cells
Enter G0 temporarily or permanently
Begin differentiation → specialized cell (e.g., neuron, RBC)
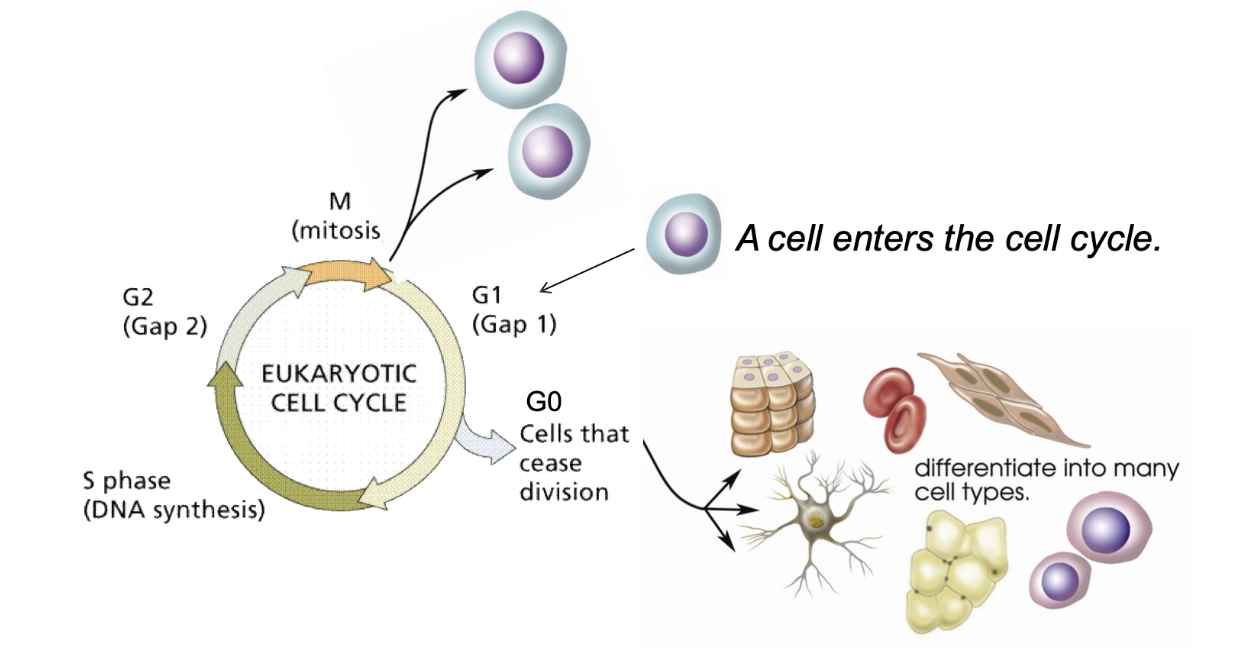
Why is balance between division and differentiation important?
Too much division → tumors
Too much differentiation → loss of regeneration
What are the phases of mitosis?
Interphase (G1, S, G2): Prepares for mitosis
Prophase: Chromosomes condense, centrosomes separate. mitotic spindle assembles
Prometaphase: Chromosomes attach to spindle via kinetochores
Metaphase: Bipolar attachment, chromosomes align at equator
Anaphase: Sister chromatids pulled apart
Telophase: Chromosomes decondense, nuclear structures reform, mitotic spindles disassemble
Cytokinesis: Cell membrane pinches → two daughter cells
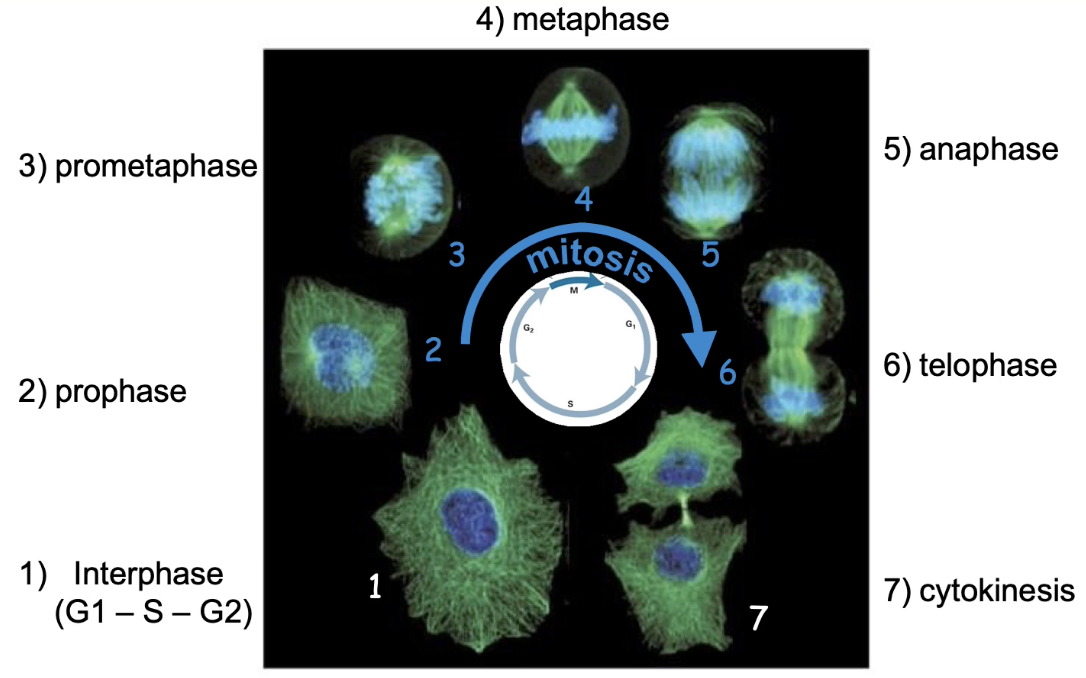
What labels are used in imaging mitosis?
DAPI = DNA (blue)
Anti-beta-tubulin antibody = spindle microtubules (green)
What are the key events during mitosis shown in the animation?
Prophase: Chromosomes condense → 4n; centrosomes move to poles
Prometaphase: Nuclear envelope breaks down; kinetochores attach to microtubules
Metaphase: Chromosomes align at equator via kinetochore microtubules
Anaphase: Sister chromatids pulled to opposite poles
Telophase: Nuclear membrane reforms; chromosomes decondense
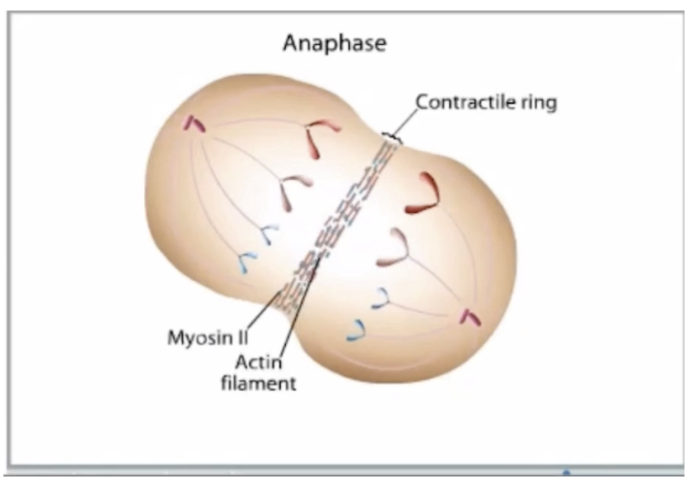
Cytokinesis: Contractile ring (actin + myosin II) → 2 identical daughter cells (2n)
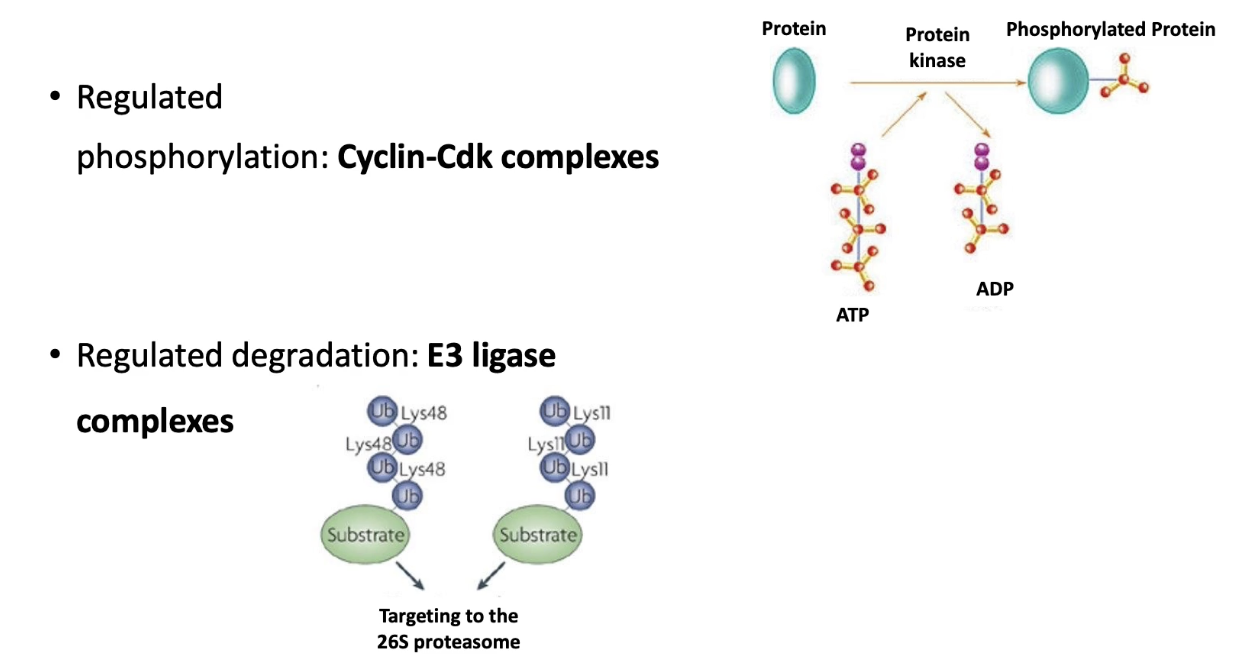
What two classes of proteins regulate the sequence of mitosis?
Cyclin-dependent kinases (CDKs):
Heterodimers (CDK + cyclin)
Phosphorylate target proteins → trigger cell cycle events
E3 ubiquitin ligases:
Target proteins for degradation via proteasome
Remove cyclins or inhibitors at key checkpoints
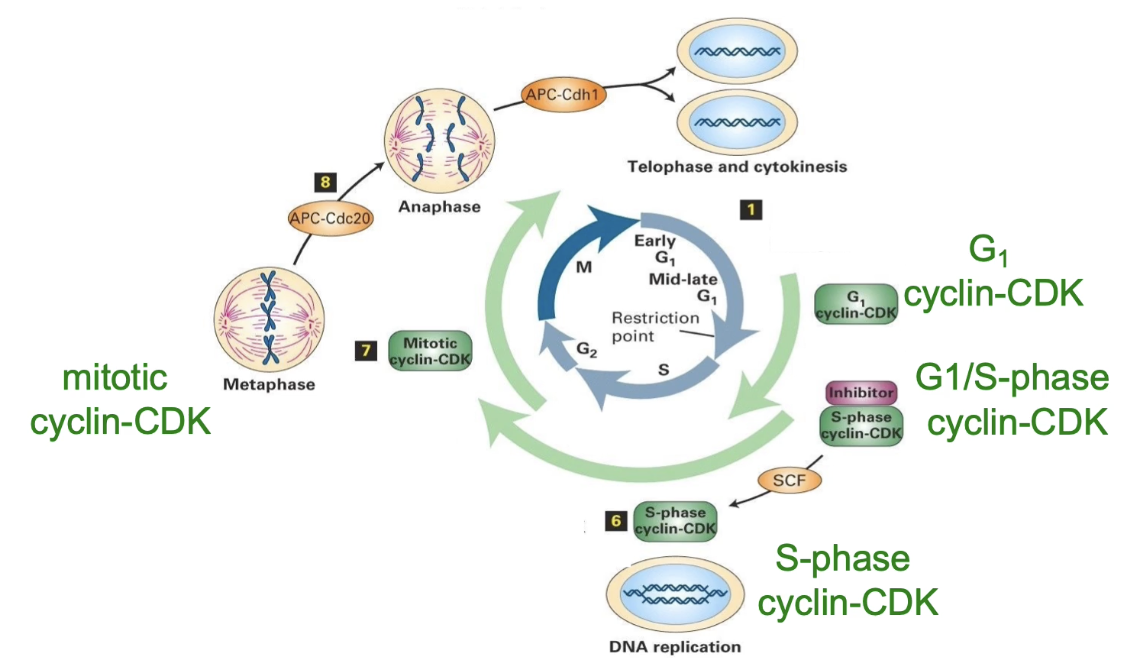
What are the four major Cyclin-CDK complexes in the cell cycle?
G1 Cyclin-CDK: Active in G1 → prepares for S phase
G1/S Cyclin-CDK: Transitions G1 to S phase
S-phase Cyclin-CDK: Initiates DNA replication
Mitotic Cyclin-CDK: Regulates prophase & mitotic changes
What is common across all Cyclin-CDK complexes?
Same structure
Same kinase activity
Differ in targets and timing
What are the 3 major E3 ligase complexes and their functions?
SCF complex:
Promotes G1 to S phase transition
APC-Cdc20:
Regulates metaphase to anaphase transition
APC-Cdh1:
Mediates exit from mitosis
APC (anaphase promoting complex)
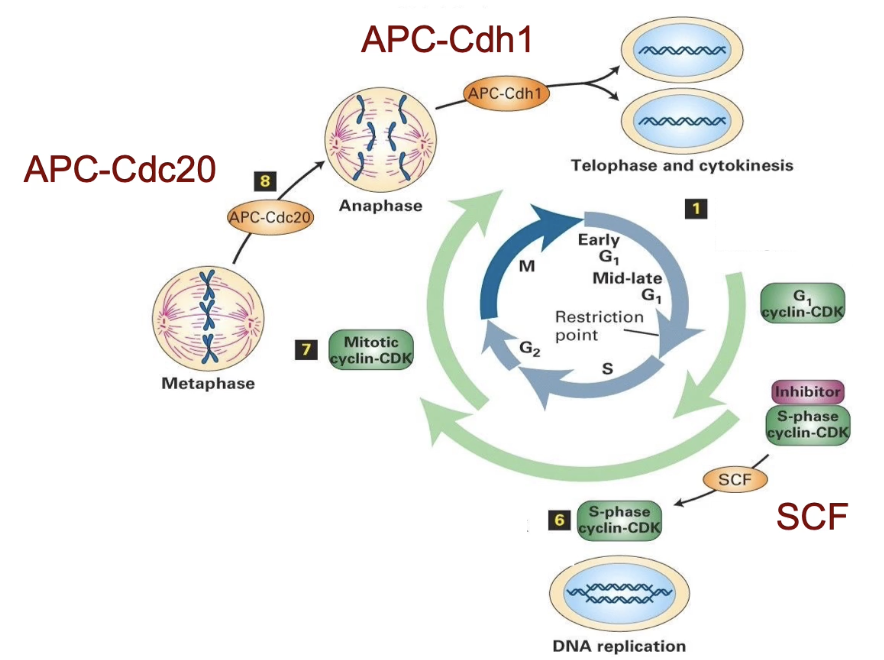
What do Cdc20 and Cdh1 do?
Accessory proteins that determine APC’s target specificity
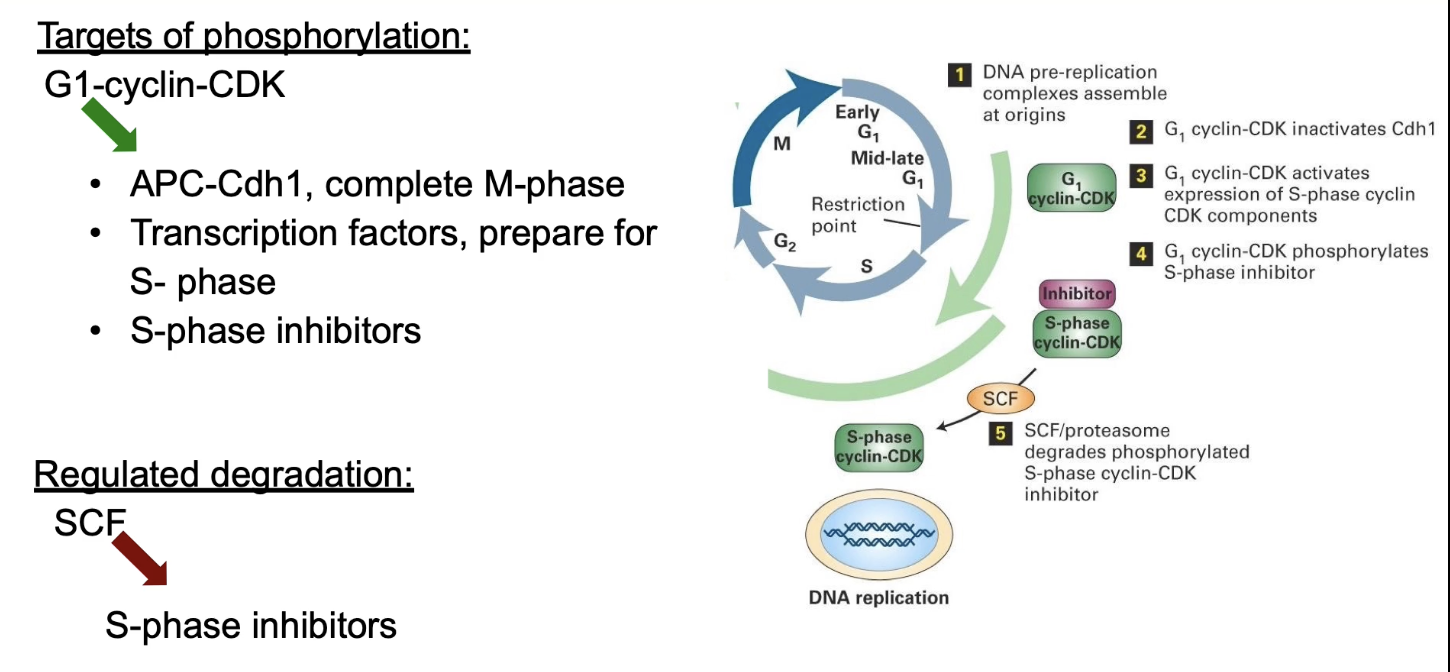
What are the 3 main targets of G1 Cyclin-CDK?
APC-Cdh1:
Phosphorylation signals mitosis completion
Transcription factors:
Activated to express S-phase genes (e.g., DNA polymerase)
S-phase inhibitors:
Phosphorylation → target for SCF ligase → degraded → S-CDK activated → initiates DNA replication
What are the functions of G1/S Cyclin-CDK?
Activating mitosis related genes
Activates transcription of mitotic genes (e.g., M-phase cyclins)
Preparing centrosomes
Phosphorylates proteins involved in centrosome replication, an important part of forming the mitotic spindle

What are the functions of S-phase Cyclin-CDK?
Triggering DNA replication
Activates pre-replication complex at origins of replication on the DNA
Preventing over-replication
Phosphorylated proteins to ensure that each origin fires only once per cycle
Delaying mitosis if DNA isn’t ready
Phosphorylates M-phase CDK to inhibit it until DNA replication is complete
What does M-phase Cyclin-CDK phosphorylate during prophase?
Chromosomal proteins → condensation
Nuclear lamins → envelope breakdown
MAPs → spindle assembly
Kinetochore proteins → chromosome-spindle binding
APC complex → mitotic progression
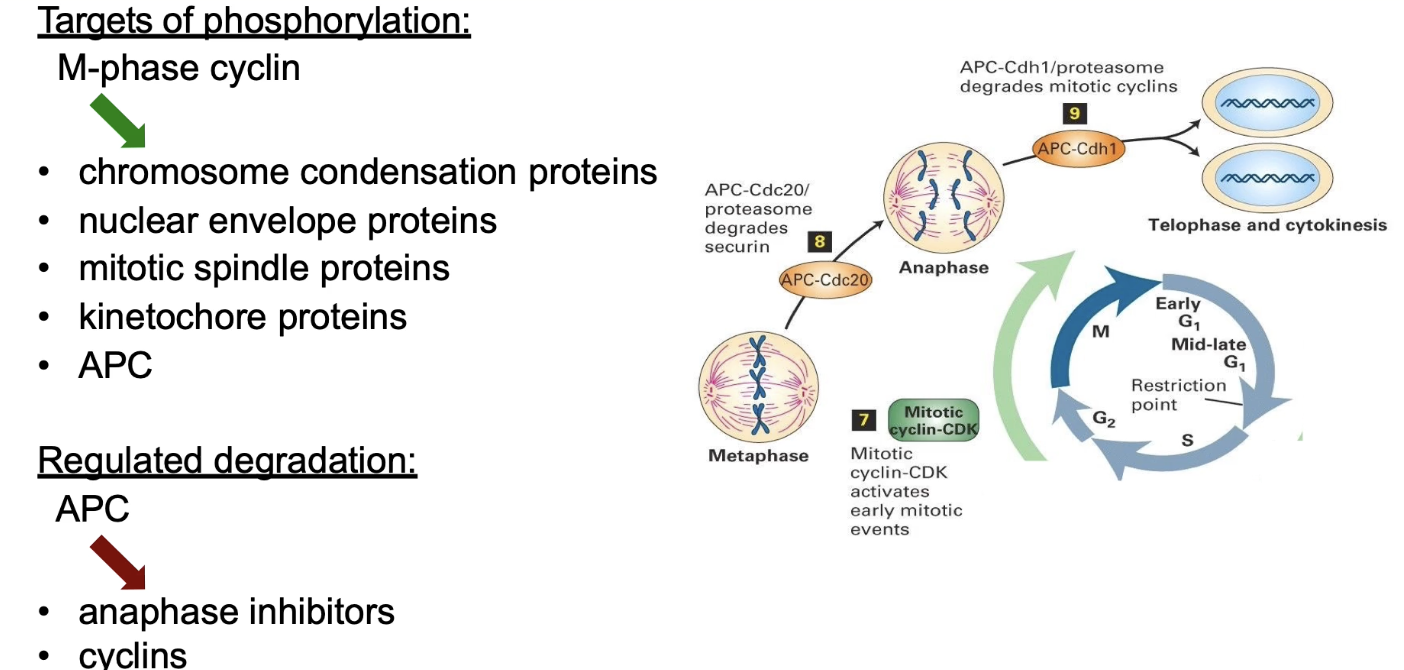
When does protein ubiquitination/degradation occur in mitosis?
Anaphase onset: Anaphase inhibitors degraded
Mitotic exit: Mitotic cyclins degraded via MEN (mitotic exist network)
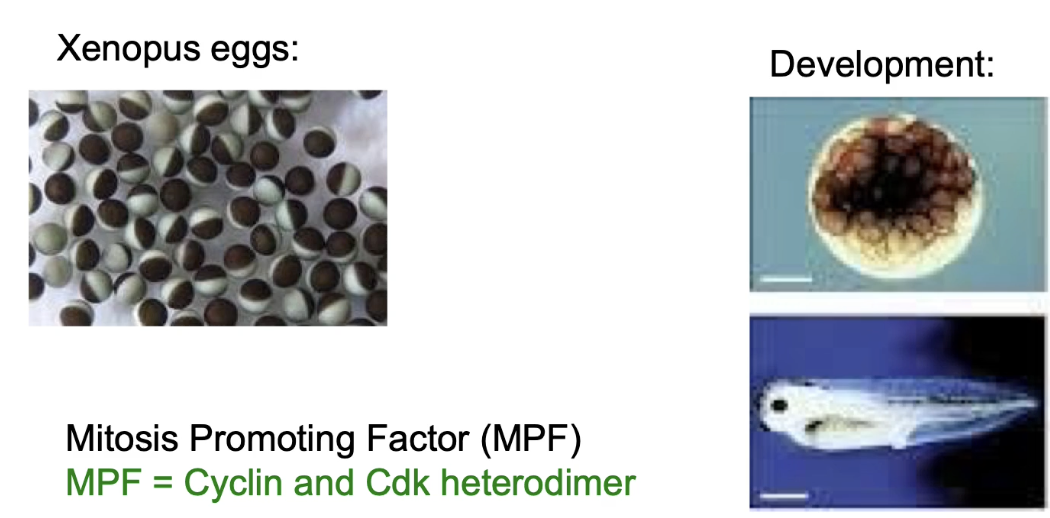
What is MPF and how was it discovered?
MPF (Maturation/Mitosis Promoting Factor) discovered in frog eggs (Xenopus)
Induces meiosis completion & 11 mitotic divisions → forms blastocyst → further cell division + differentiation into tadpole
Identified by Masui & Markert, 1971
Later shown to be M-phase Cyclin-CDK
Why use synchronized embryos in cell cycle research?
High synchrony makes it easier to isolate and study cell cycle regulators biochemically
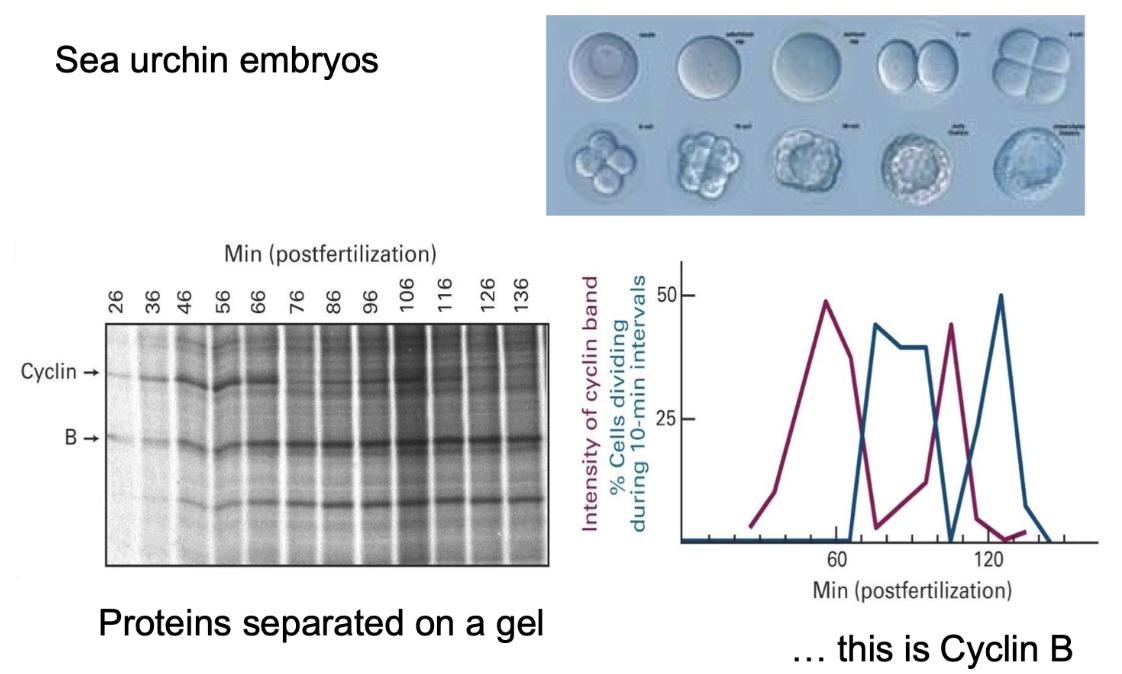
What did Tim Hunt and Joan Ruderman discover using sea urchin embryos?
Identified cyclins: proteins with cyclic synthesis/degradation
Used radiolabeled proteins and gel electrophoresis
Observed Cyclin B levels oscillate with mitotic activity
Graph:
Pink line = how much Cyclin B was present over time.
Blue line = how many cells were in mitosis at that time.
Correlation between cyclin levels and mitosis:
↑ Cyclin B → ↑ cells in mitosis
↓ Cyclin B → ↓ mitotic cells
Showed Cyclin B regulates M-phase Cyclin-CDK activity and directly correlates with whether or not a cell is dividing.
What did live imaging of Cyclin B in HeLa (human) cells show?
Cyclin B present during interphase & early mitosis
Rapid drop in Cyclin B during anaphase
This drop in Cyclin B is a key signal for the cell to exit mitosis and finish division
Why was it surprising that cyclin regulates the cell cycle?
Cyclin has no enzymatic activity, unlike MPF (a kinase)
Big Question: How could a non-enzymatic protein like Cyclin B control something as important as the cell cycle?
What did Andrew Murray’s in vitro experiment involve?
Used egg extracts (mRNA + proteins for cell division)
Measured:
MPF activity - via histone H1 phosphorylation
Cyclin B levels - via gel
Mitotic behaviors - observed by adding sperm nuclei into the extract to see if they behaved like they do in real cells (e.g., chromosome condensation, nuclear envelope breakdown)
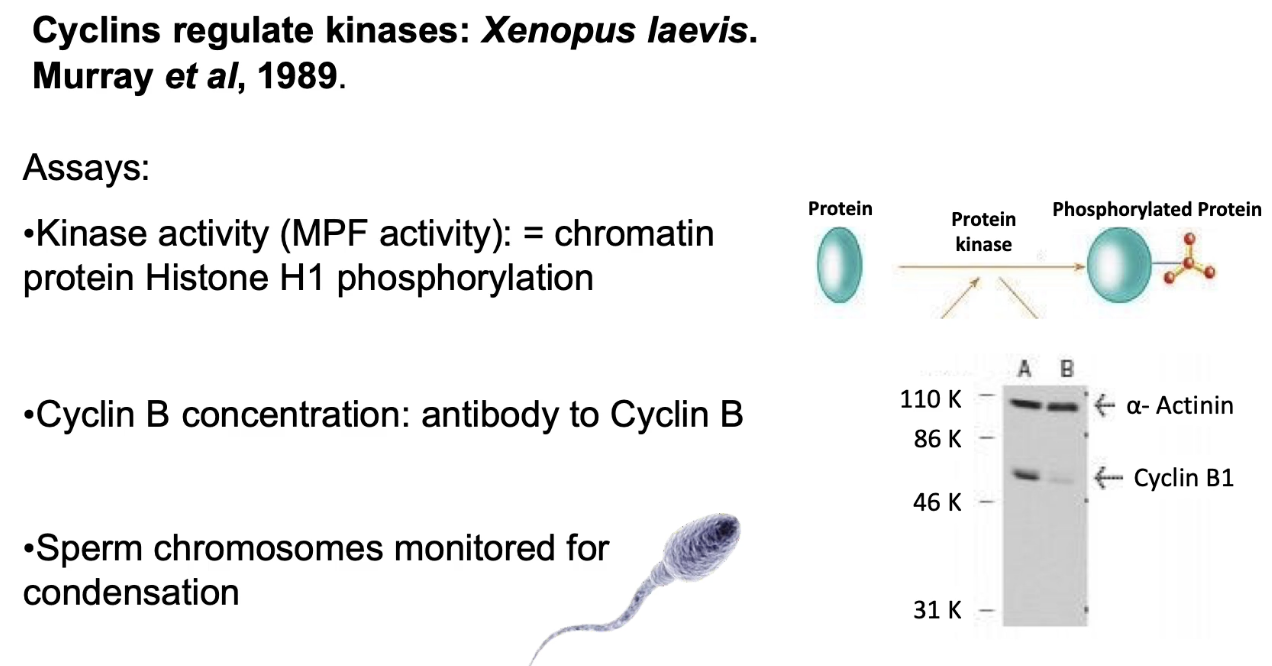
What happened when sperm nuclei were added to egg extracts?
Cyclic mitotic behaviors occurred:
Condensation of chromosomes and breakdown of the nuclear envelope (typical of early mitosis).
Decondensation and reformation of the envelope (typical of late mitosis).
Synchronized with ↑/↓ Cyclin B and MPF activity:
↑ Cyclin B levels = ↑ MPF activity increased → mitosis started.
↓ Cyclin B levels dropped = ↓ MPF activity decreased → cells exited mitosis.
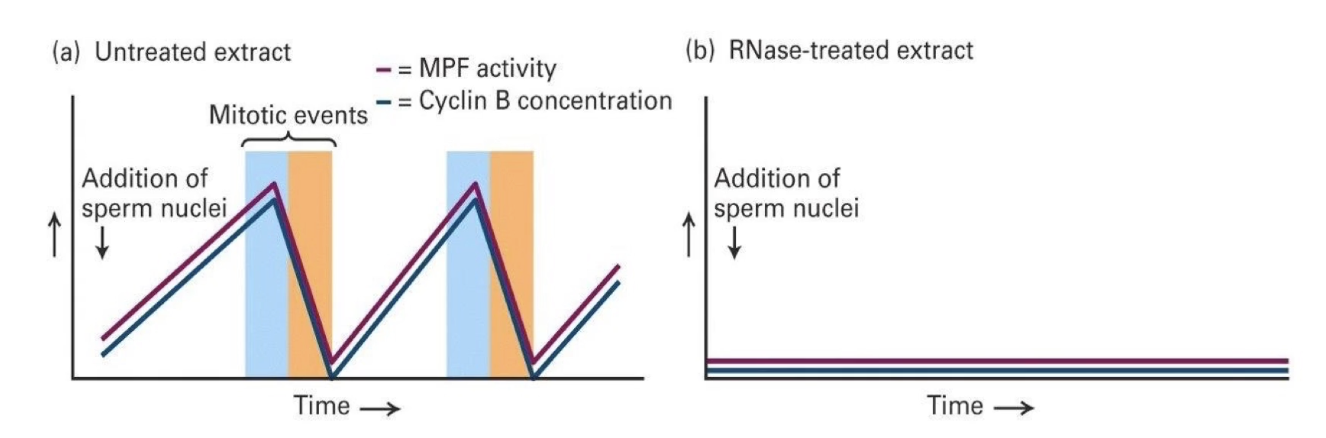
What did RNase-treated extract experiments show?
No mRNA (but tRNA and sRNA for protein synthesis intact) → No new Cyclin B made → No MPF activity
Conclusion: Cyclin B is necessary for MPF activation and mitosis
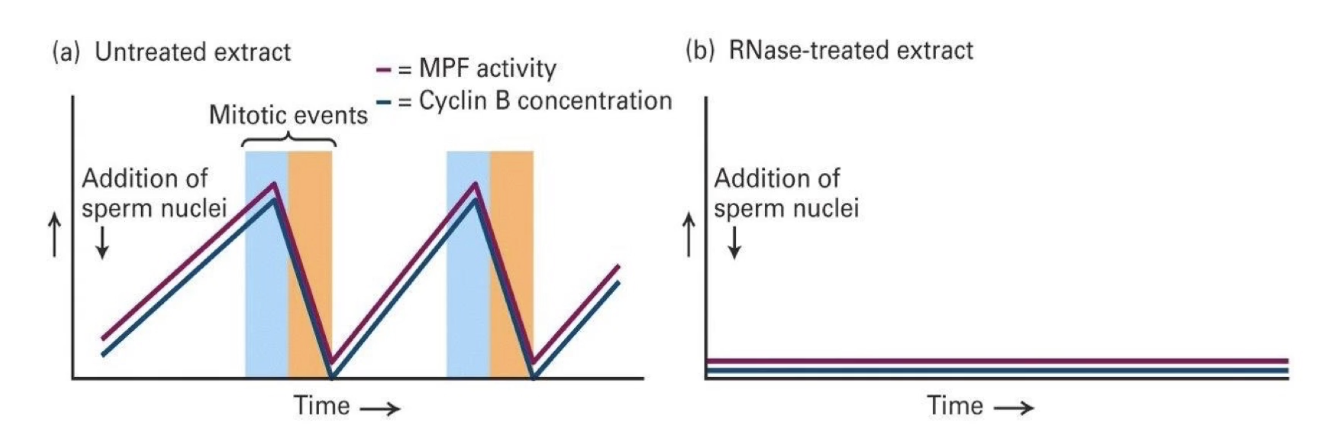
What happens when Cyclin B mRNA is added to RNase-treated extract?
Restores synchronized cycling behaviors
Only Cyclin B is synthesized
Cycling CDK activity and mitosis behaviors return
Conclusion: Cyclin B is sufficient to restart the cycle, even if it’s the only new protein made.
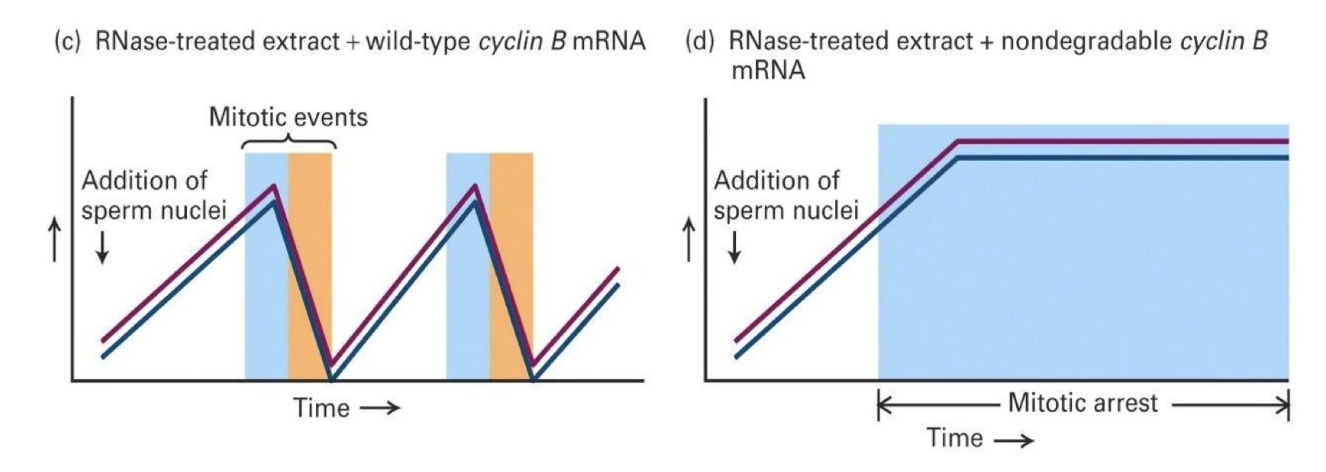
What happens when nondegradable Cyclin B mRNA is added?
Cyclin B levels stay high
CDK activity remains high
Mitotic arrest occurs
The chromosomes condensed but never decondensed, and mitosis didn’t finish
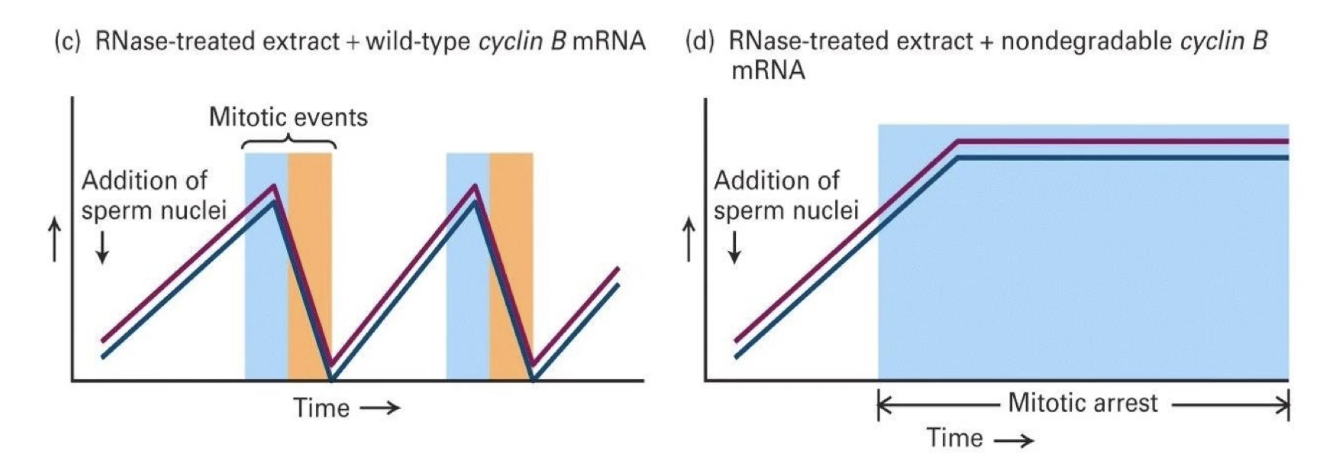
What do these experiments show?
Cyclin B is necessary for CDK activation
Its degradation is needed to complete mitosis
What did microscopy results show for degradable vs. nondegradable Cyclin B mRNA?
Degradable:
Chromosomes: anaphase → telophase
Spindle: assembles/disassembles
Non-degradable:
Chromosomes: undergo anaphase but fail to decondense
Spindle: fails to disassemble
Conclusion: Cyclin B degradation is required to exit mitosis

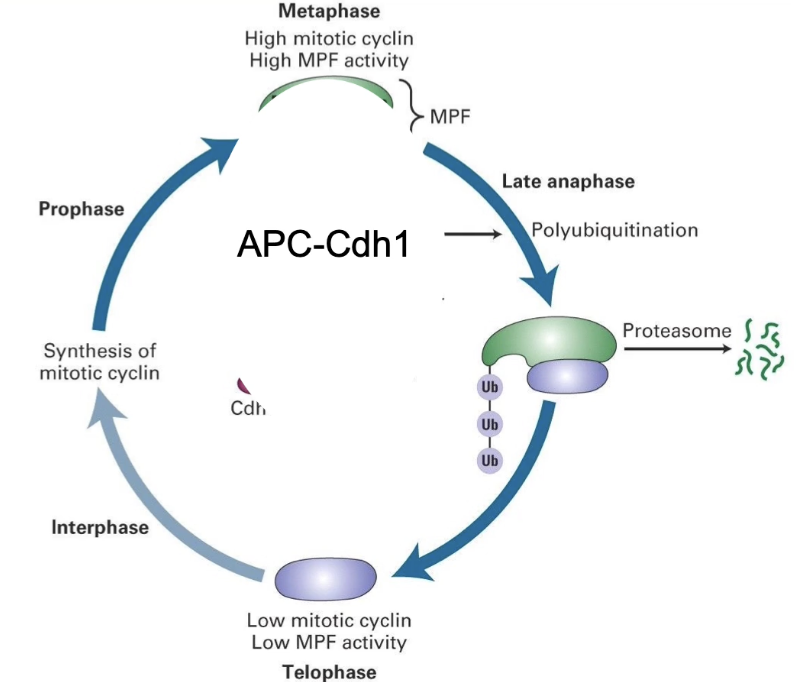
What complexes degrade Cyclin B and the outcome?
APC-Cdc20: Activated at anaphase, it begins Cyclin B degradation.
APC-Cdh1: Takes over after anaphase to finish degrading Cyclin B, allowing the cell to exit mitosis and reset.
Outcome:
CDK inactivation
Cell exits mitosis
What sequence allows APC-Cdc20 to recognize Cyclin B and its significance?
Destruction box (D-box) near N-terminus: RxxLxxxxN/Q
Position 1: Arginine
Position 4: Leucine
Position 9: Asparagine/Glutamine
Significance:
Necessary: mutations prevent degradation
Sufficient: adding it to another protein (GFP) causes cyclical degradation

What happens when D-box peptide is added to in-vitro extracts?
Low amounts of D-box peptide caused delays in mitosis.
High amounts completely blocked cells in metaphase—they couldn’t move into anaphase.
Why?
Because APC was so busy binding the excess D-box peptides, it couldn't bind and degrade a real protein needed to progress.
This suggested APC has a second critical target: an anaphase inhibitor.

What is the anaphase inhibitor?
Securin
Sister chromatids are held together by a cohesin complex (Smc1, Smc3, Scc1).
A protein called separase can cut Scc1 to start chromatid separation—but it’s kept inactive by securin.
When APC-Cdc20 is activated, it degrades securin.
This frees separase, which cuts Scc1, a key component of the cohesin ring.
Once cut, the chromatids separate and anaphase begins.
So, APC-Cdc20 is essential to initiate anaphase by removing the block (securin) on chromosome separation.
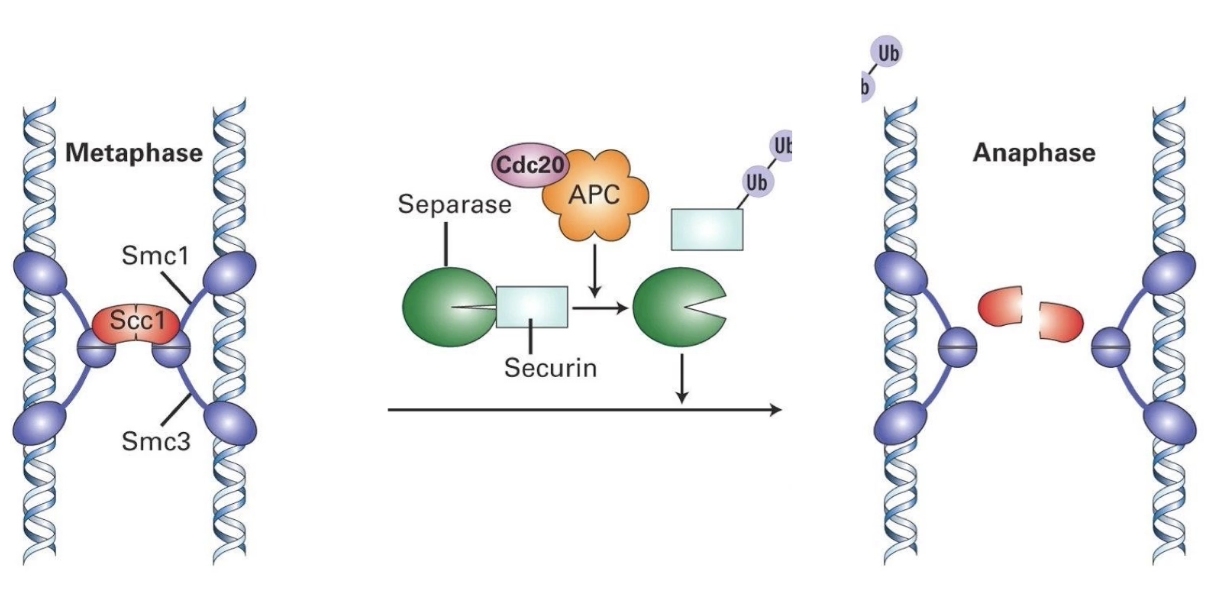
How is securin targeted for degradation?
By APC-Cdc20
Cdc20 acts as specificity factor for APC
Additional note
APC subunit is phosphorylated by CyclinB-CDK, prepping for anaphase
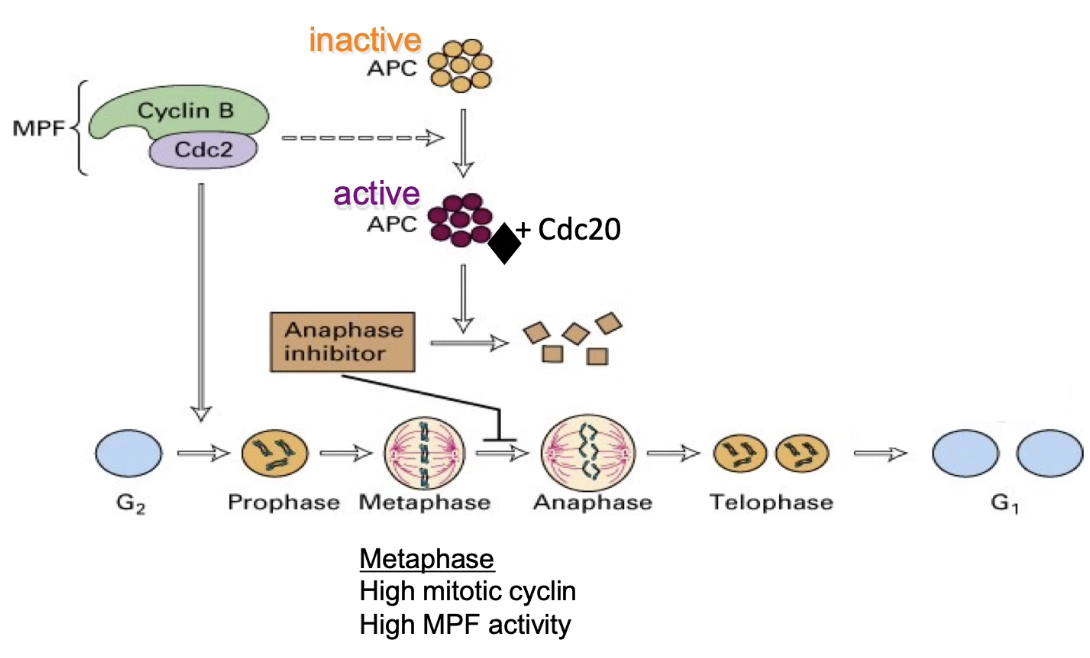
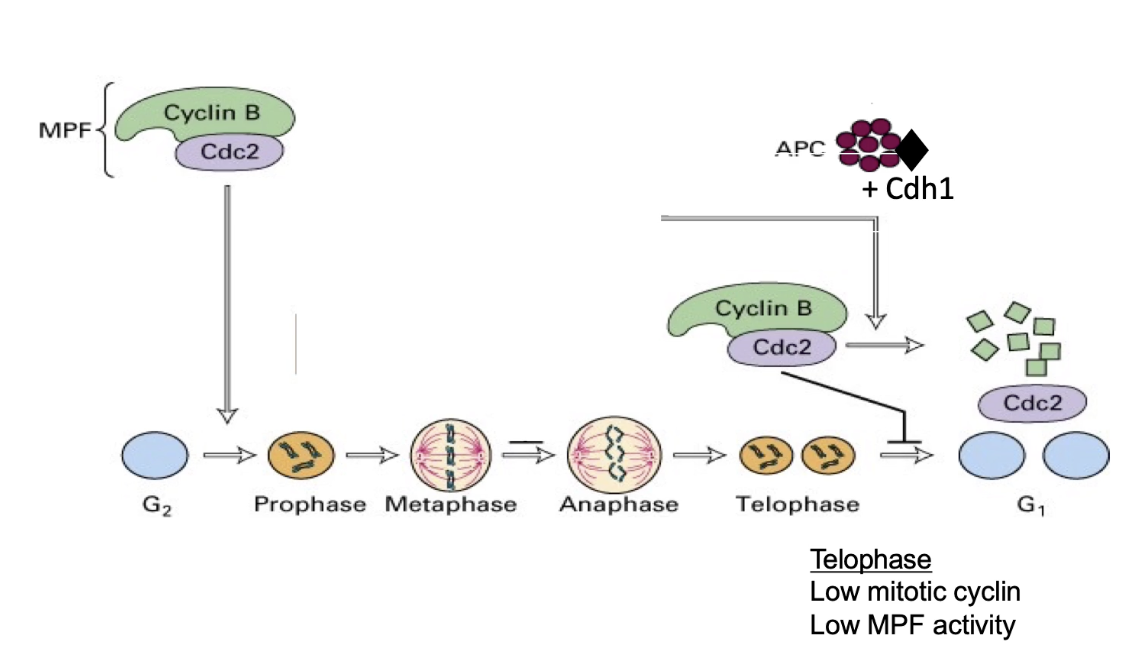
How is Cyclin B degraded at mitotic end?
1. APC switches specificity:
After anaphase, APC changes its regulatory subunit from Cdc20 → Cdh1.
2. APC-Cdh1 targets Cyclin B:
During telophase, APC-Cdh1 ubiquitinates Cyclin B, marking it for proteasomal degradation.
3. Inactivation of CDK:
Cyclin B degradation → inactivates CDK (M-phase kinase).
This allows the cell to exit mitosis and enter G1 phase.
4. APC deactivation in G1:
Without Cyclin B-CDK activity, phosphatases dephosphorylate APC → APC is inactivated in G1.
5. Cyclin B initiates its own end:
Interestingly, Cyclin B-CDK is required to activate APC at mitotic entry.
So, Cyclin B helps trigger the system that eventually leads to its own destruction.
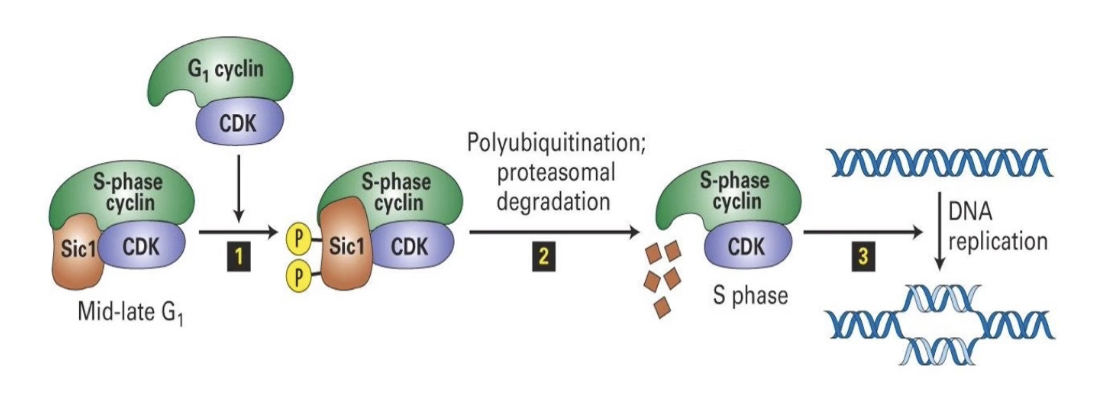
What is SCF and its function?
E3 Ligase SCF = Skp, Cullin, F-box protein complex
Active in mid-G1 phase
Targets S-phase inhibitor Sic1
Sic1 inhibits S-phase CDK until the cell is ready
G1-CDK phosphorylates Sic1 → SCF recognizes and ubiquitinates Sic1 → Sic1 degraded by proteasome
S-phase CDK is activated → cell enters S-phase
Ensures irreversible, one-way progression through cell cycle
What regulates progression through the cell cycle?
CDK-cyclin complexes → phosphorylation
E3 ligases (APC, SCF) → degradation of regulators
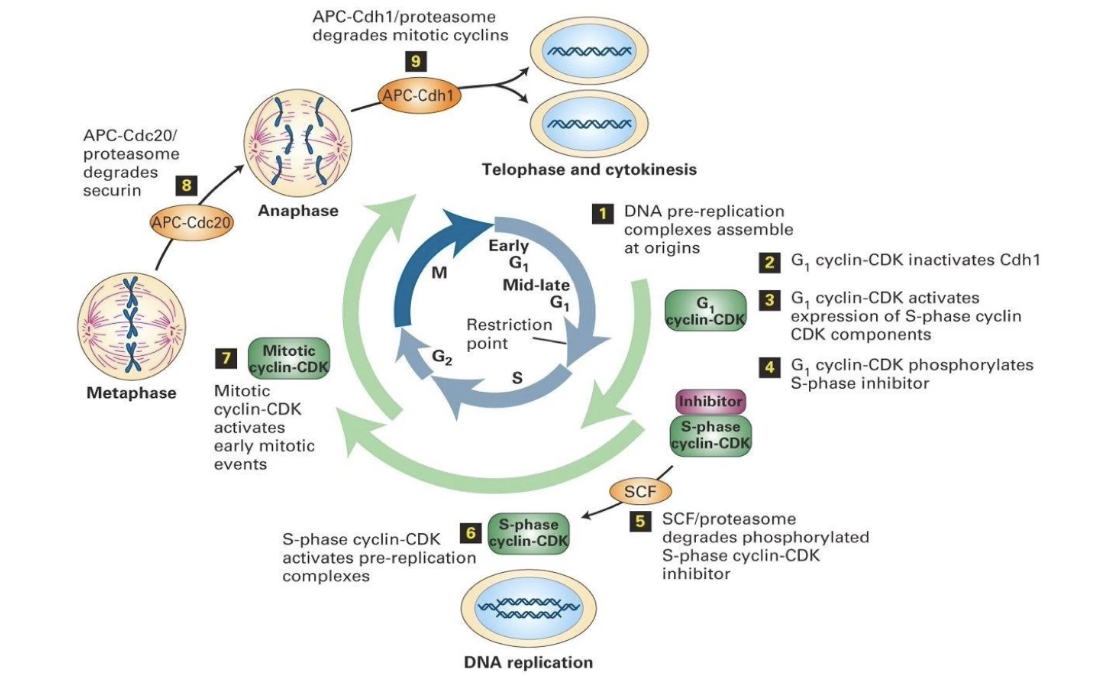
Animation Summary
Early G1:
DNA prereplication complexes dephosphorylated
Assemble at replication origins
Late G1:
G1-CDKs synthesized → activate transcription factors
Induce expression of S-phase CDK components
S-phase CDK blocked by inhibitor (e.g., Sic1)
Start of S phase:
G1-CDK phosphorylates inhibitor → degradation
S-phase CDK activated
Triggers DNA replication (1 round only)
Cohesins hold sister chromatids together
S phase & G2:
Mitotic CDKs produced, but kept inactive
M phase:
Mitotic CDKs activated → initiate mitosis
APC activated → degrades cohesin regulators
Allows chromatid separation (anaphase)
End of M phase:
APC degrades mitotic CDKs
Cytokinesis completes → new cell cycle starts
How are novel cell cycle regulators identified?
Use of genetic screens (unbiased approach)
Random mutations created across genome
Screen for cell cycle phenotypes:
Inhibited division
Excessive division
What is a temperature-sensitive (TS) mutation and how is it used?
TS mutation = protein functional at low temp (24°C), misfolds at high temp (37°C)
Enables on/off control of protein function
Mutant cells: grow at 24°C, but not at 37°C
Wild-type cells: grow at both temperatures
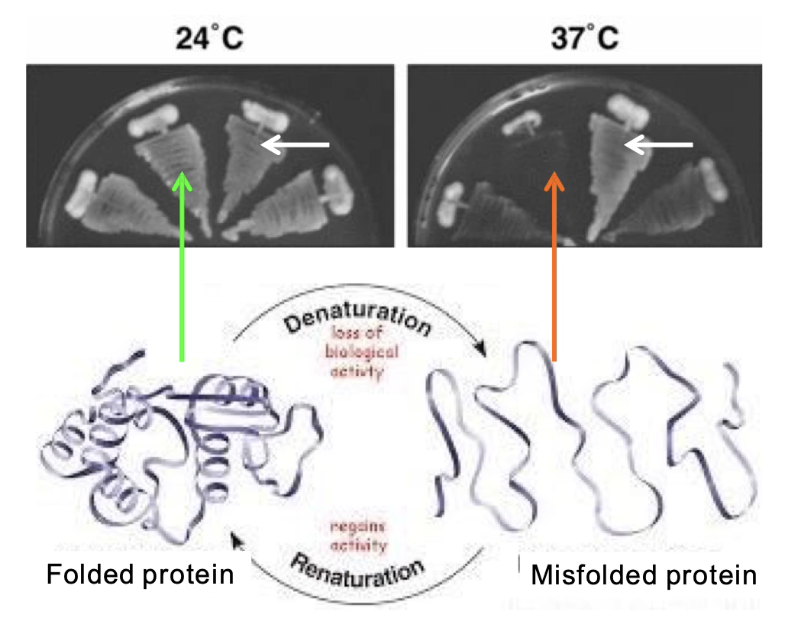
What is Schizosaccharomyces pombe and why is it used?
A model organism: fission yeast (S. pombe)
Elongated, rod-shaped cells
Used to study cell cycle and division mechanisms
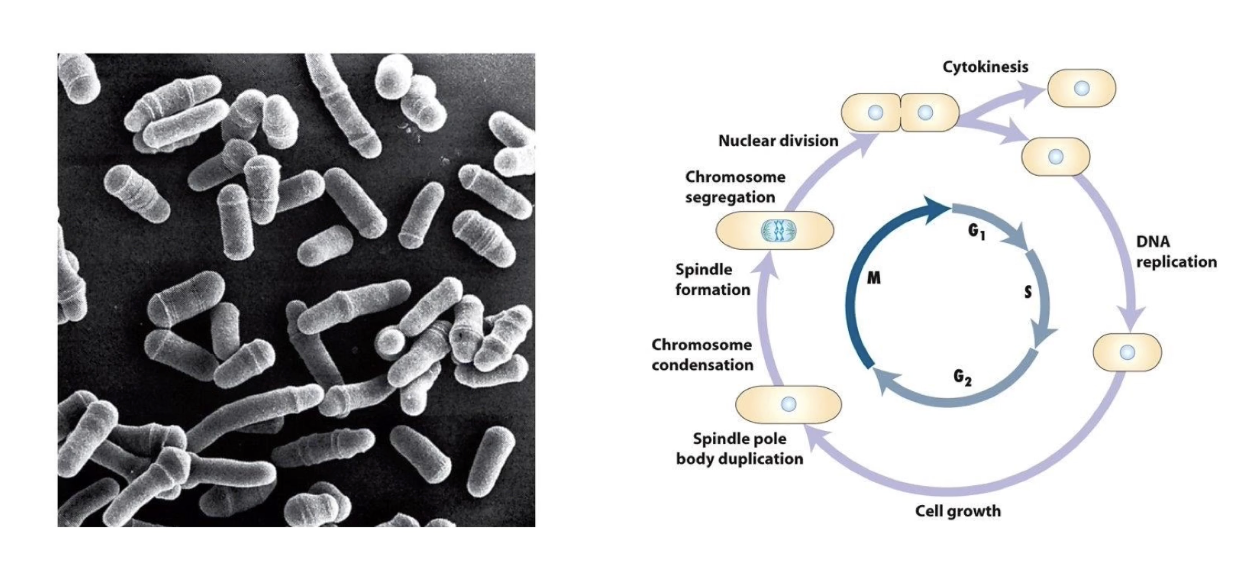
What does the movie of fission yeast show?
Fluorescent DNA labeling
Shows nuclear division → followed by cytokinesis
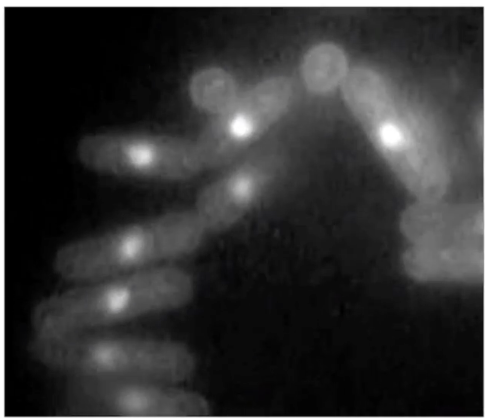
What are the two main phenotypes of cdc mutants?
Elongated phenotype: G2 delay, continued growth
Wee phenotype: Early mitosis entry, smaller cells
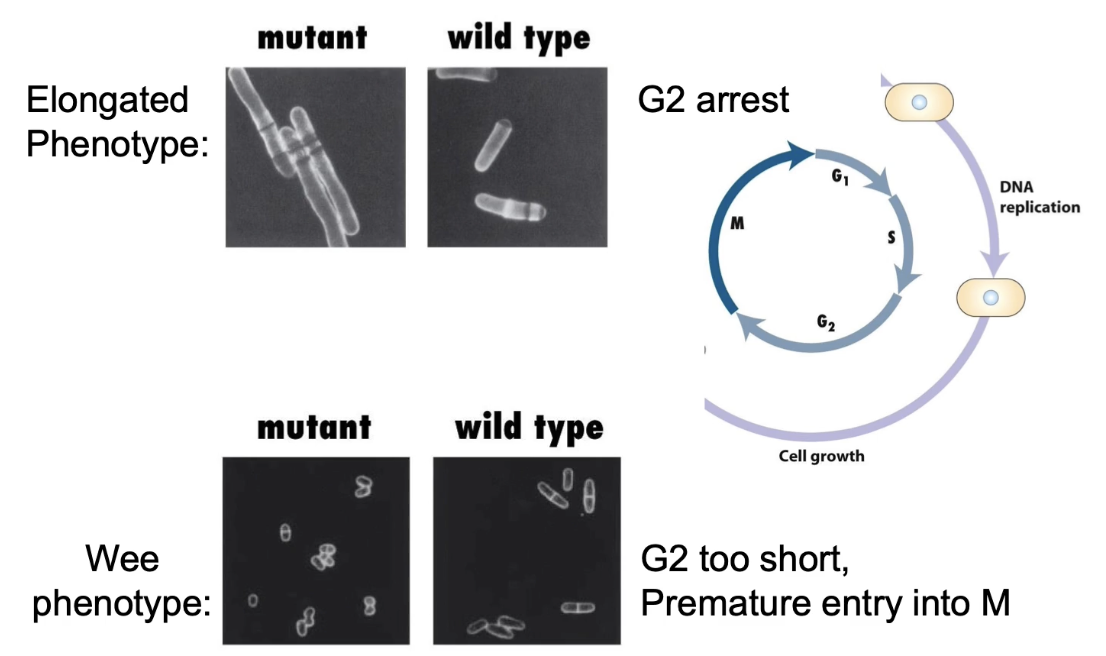
What do different cdc2 mutations cause?
Wild-type (cdc2⁺) → normal division
Loss-of-function (cdc2⁻) → elongated phenotype
Gain-of-function/dominant (cdc2ᴰ) → wee phenotype
What is the role of Cdc2 protein in the cell cycle?
Cdc2 promotes mitotic entry and cell division
Loss → no division (elongated)
Gain → early/frequent division (wee)
Cdc2 = CDK of MPF
Is a 34 kDa protein with kinase activity, forms heterodimer with Cdc13 cyclin
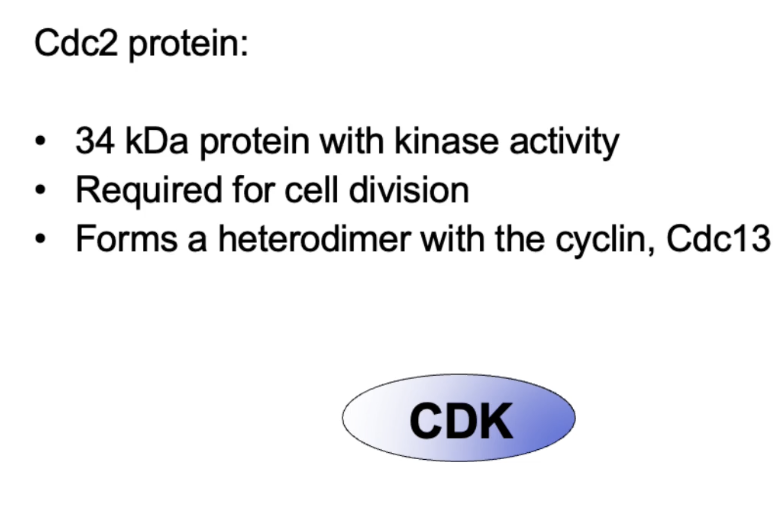
What is the role of Cdc13 in S. pombe?
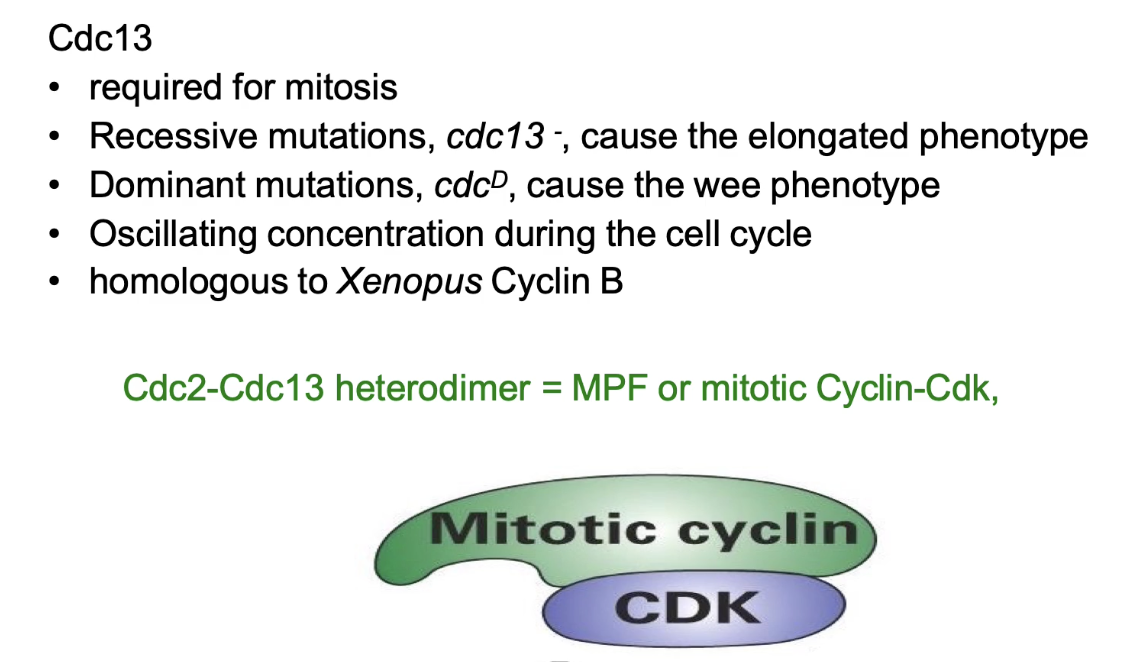
Regulates MPF activity
Loss → elongated; Gain → wee
Oscillating concentration during cell cycle
Cyclin B homolog (Xenopus)
Forms Cdc2–Cdc13 complex = MPF
Only one CDK (Cdc2) and one cyclin (Cdc13) in S. pombe
Functions as all CDKs (M-phase, S-phase, G-phase)
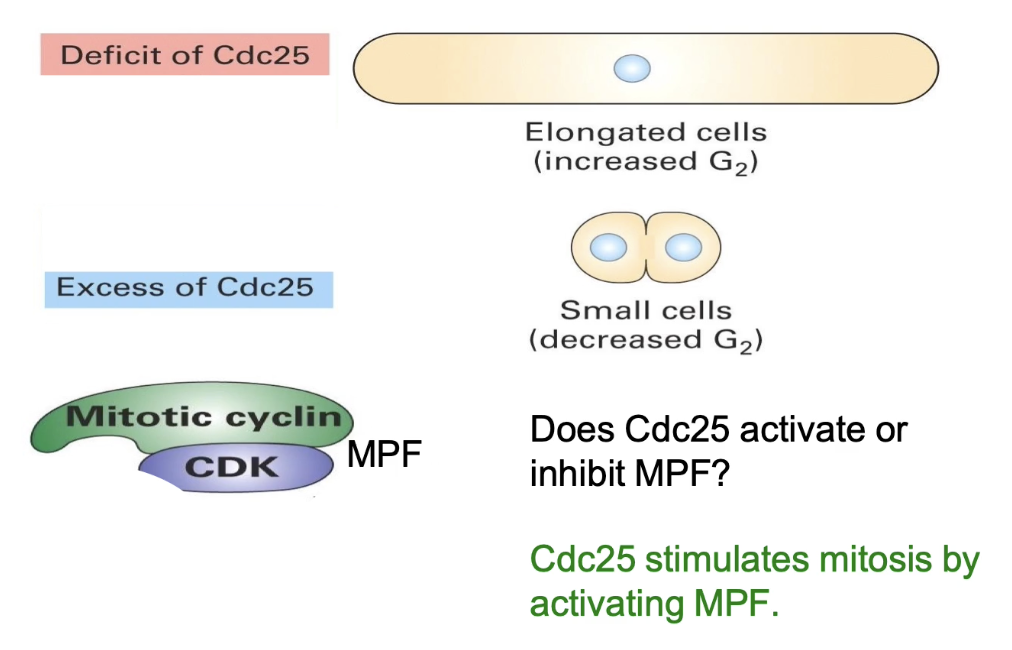
What does Cdc25 regulate, and how?
Loss (cdc25⁻) → elongated
Gain (cdc25ᴰ) → wee
Cdc25 = activator of MPF
Promotes entry into M-phase
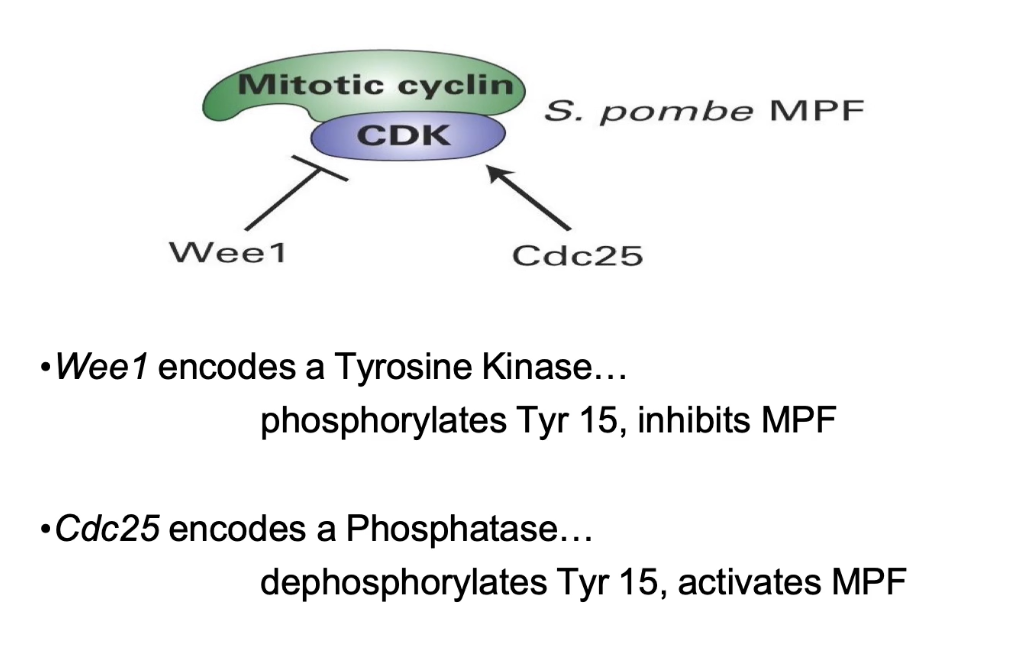
What does Wee1 regulate, and how?
Loss (wee⁻) → wee phenotype
Gain (weeᴰ) → elongated
Wee1 = inhibitor of MPF
Delays M-phase entry
Opposes Cdc25 function

What are the roles of Wee1 and Cdc25 in MPF regulation?
Wee1: Tyrosine kinase → adds inhibitory phosphate (Tyr 15 on Cdc2)
Cdc25: Phosphatase → removes Tyr 15 phosphate → activates MPF
Balance between Wee1 and Cdc25 controls mitotic entry
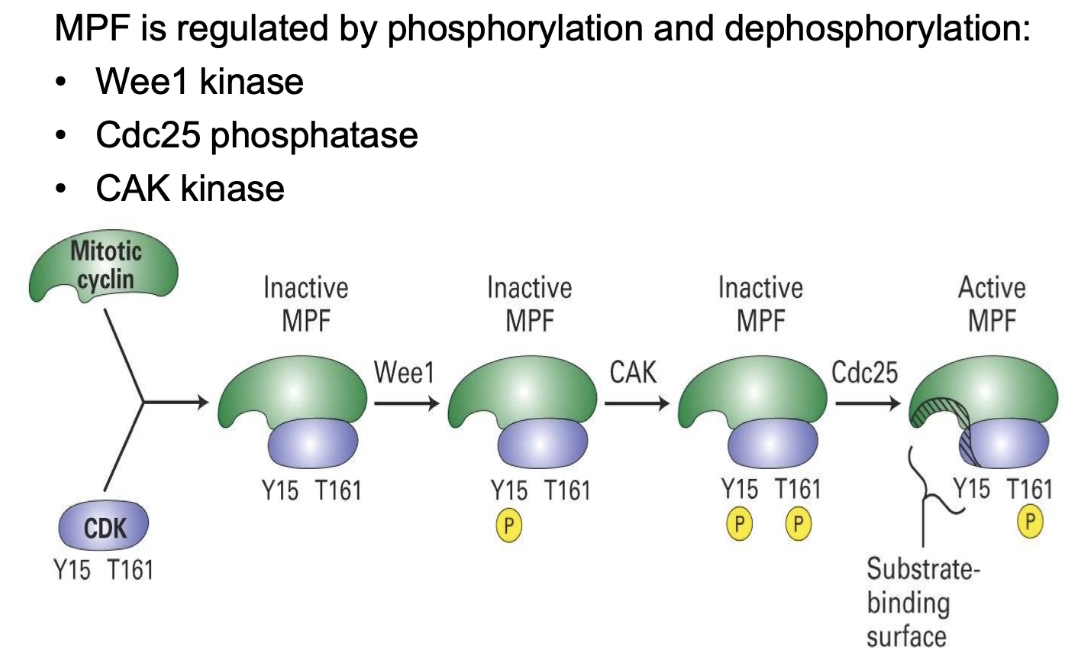
How is MPF activity regulated by phosphorylation?
Cyclin binding to CDK (Cdc2) → required for activity
Wee1 kinase → phosphorylates Y15 (inhibitory) → MPF off
CAK kinase → phosphorylates T161 (activating) → not enough alone
Cdc25 phosphatase → removes phosphate from Y15 → MPF fully activated
Double mutant (no Wee1 & Cdc25) → slow division shows CAK alone works, but less efficient and synchronized
Multiple regulators = tight, efficient cell cycle control
What is a key difference in budding yeast (S. cerevisiae) compared to fission yeast?
Forms daughter bud in G1, before S-phase
Same cell cycle regulators in S. cerevisiae as S. pombe
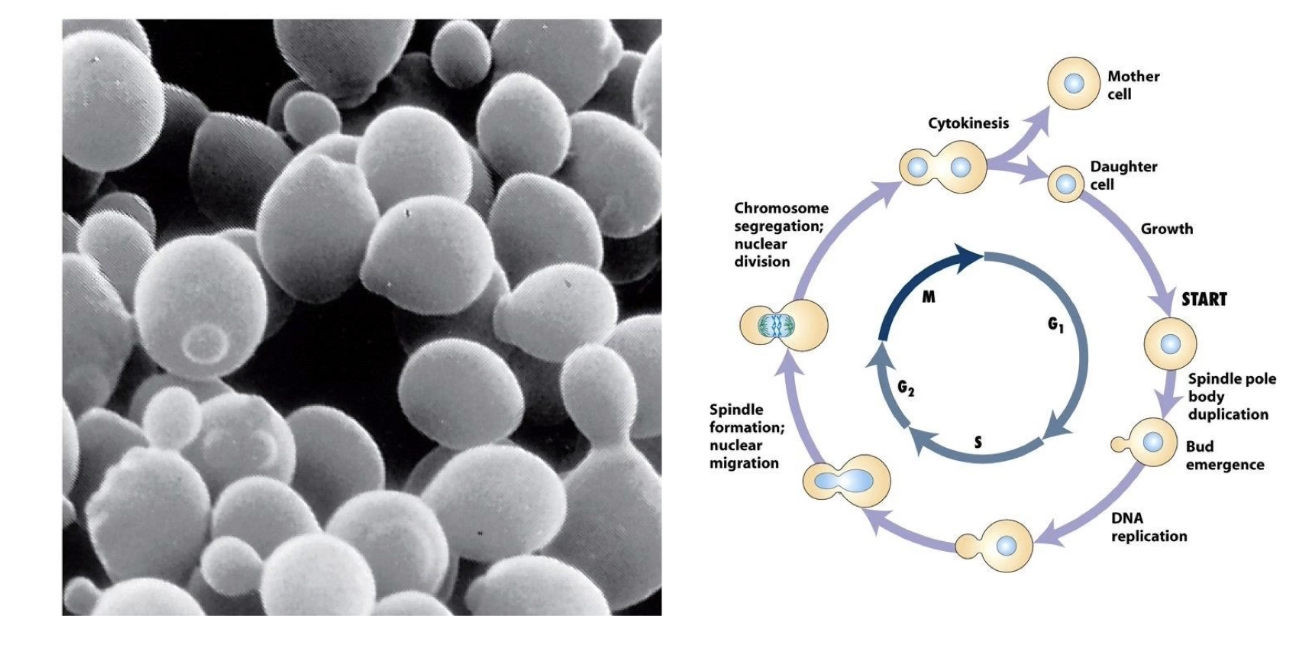
What does the Nomarski microscope movie show?
Budding yeast cells dividing
Visualizes cell morphology changes during division
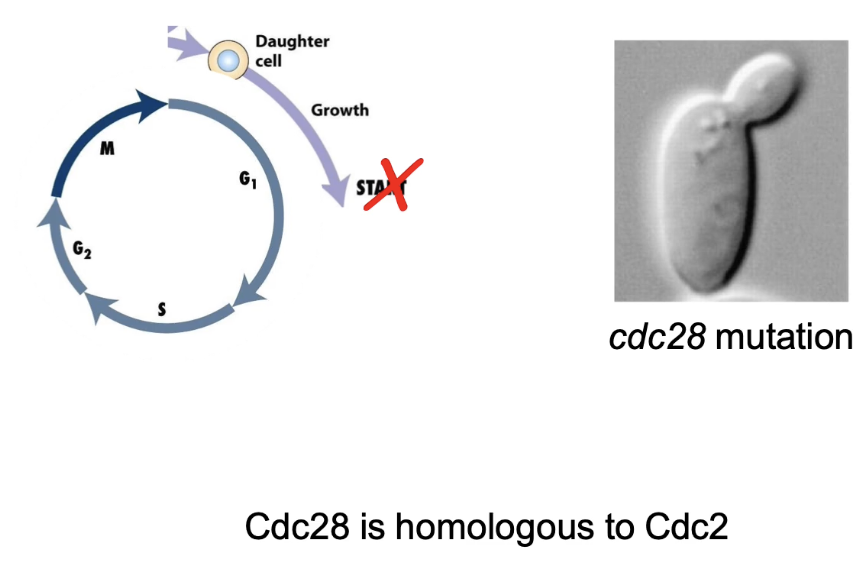
What phenotype is caused by cell cycle mutations in S. cerevisiae?
Arrest in G1
Daughter bud forms, but no S-phase
Cdc28 = homolog of Cdc2 in fission yeast
Single CDK controls cell cycle in both yeasts
Phenotypes differ slightly despite functional homology
What is functional complementation and how is it used?
Technique to identify gene that rescues mutant phenotype
Wild-type gene introduced into mutant cells
Step 1: Start with a temperature-sensitive (TS) mutant
At 25°C (permissive): cells divide normally
At 35°C (restrictive): cells arrest in G1 due to a mutation in an unknown gene
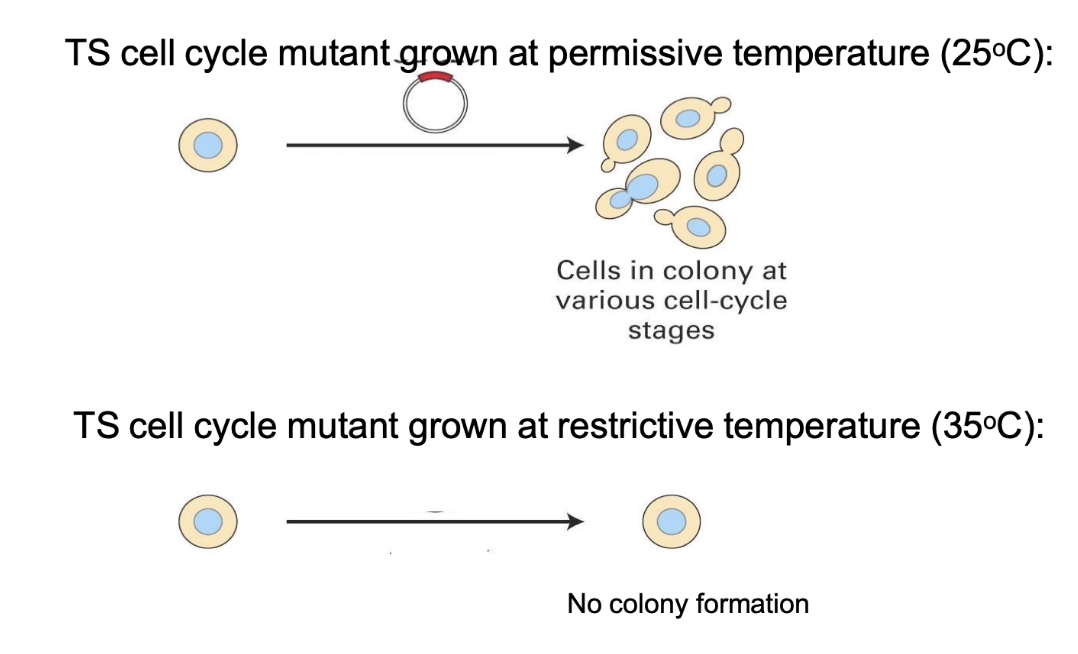
What is the purpose of using a cDNA library in functional complementation?
Step 2: Add genes from a cDNA library
A cDNA library contains DNA versions of expressed genes (no introns), made from mRNA using reverse transcriptase
This library is introduced into the mutant cells, one gene at a time, to see if any restore normal function
Screen each cDNA at restrictive temperature:
Gene X/Y → no rescue
Gene Z → rescues cell division = likely wild-type of mutated gene
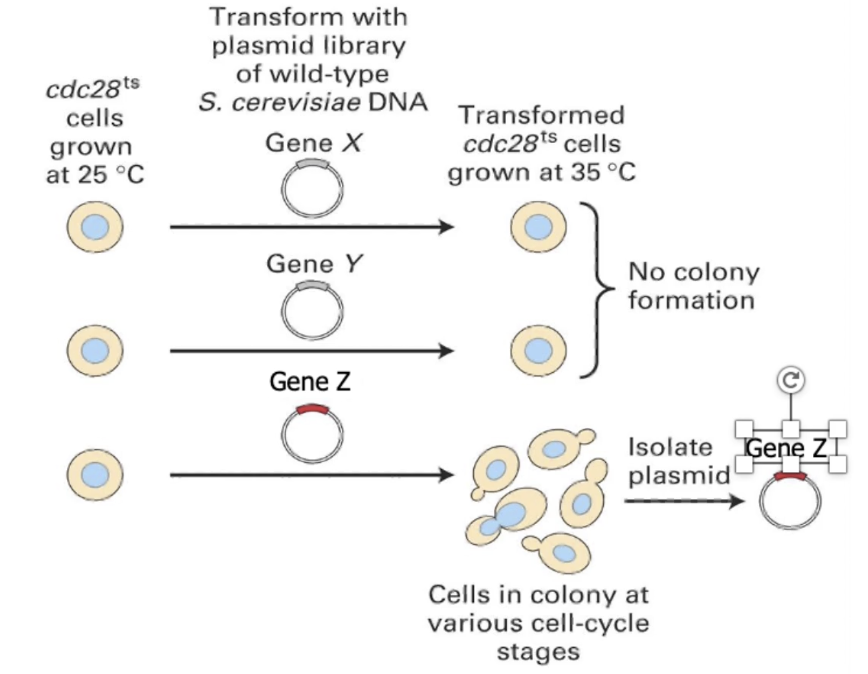
What does isolating gene Z reveal?
Step 3: Identify Gene Z
Gene Z is carried in a plasmid in bacterial cells
Extract plasmid → sequence the cDNA
Result: Gene Z = Cdc28 in budding yeast
Cdc28 codes for CDK1 protein = same as Cdc2 in fission yeast
Functional complementation shows conserved cell cycle genes across species
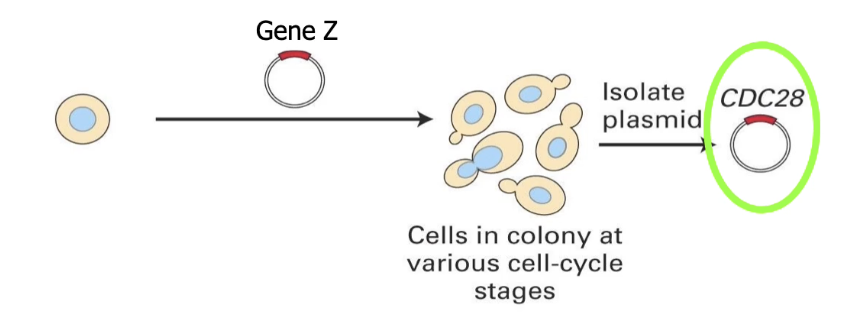
What does Cdc28 do in budding yeast?
Cdc28 = single CDK in S. cerevisiae
Binds to cyclins to regulate cell cycle
G1/S cyclins: Cln1, Cln2, Cln3 → form SPF (S-phase promoting factor)
M-phase cyclins: Clb1, Clb2 → form MPF (mitosis promoting factor)
CDK + Cyclin = active heterodimer → phosphorylates targets
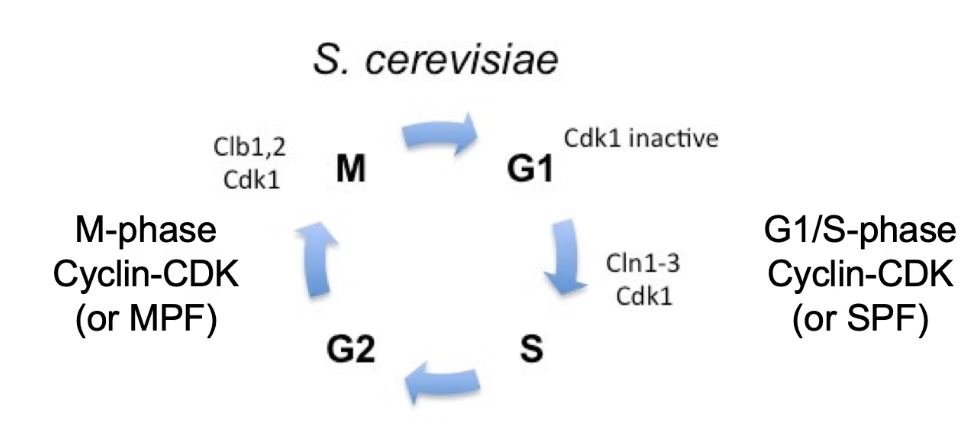
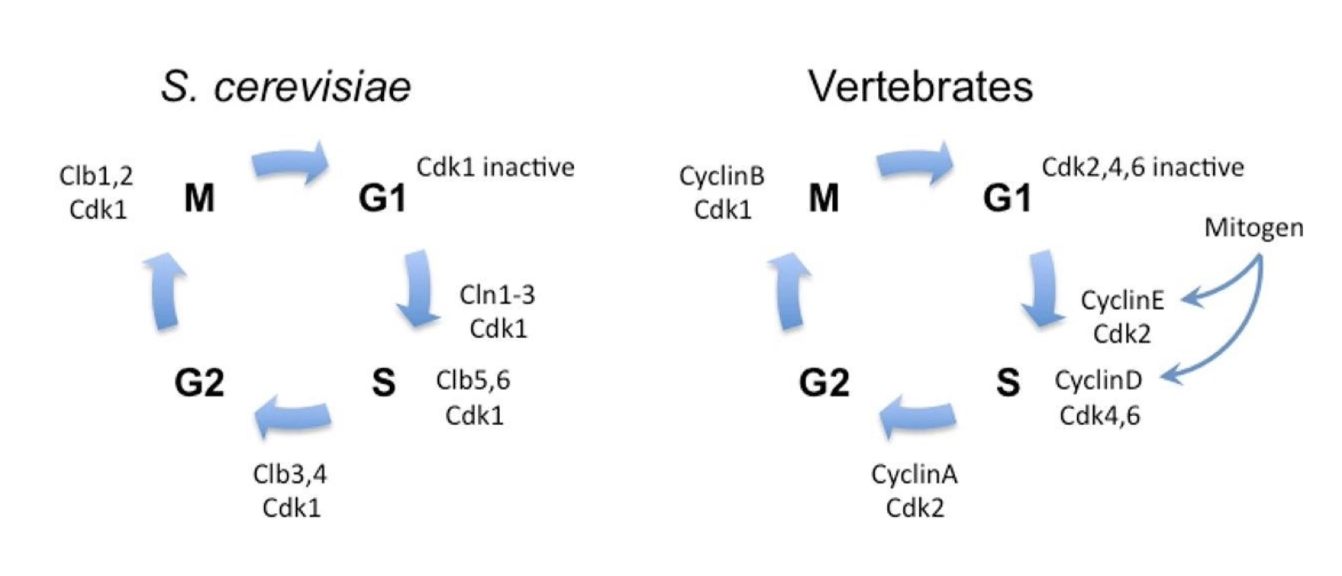
Are cell cycle regulation mechanisms conserved across species?
Yes, conserved in all eukaryotes
Cyclin-CDK complexes perform similar roles in each phase
Regulatory enzymes (kinases, phosphatases) also conserved
Vertebrates: multiple CDKs + multiple cyclins, but homologous functions
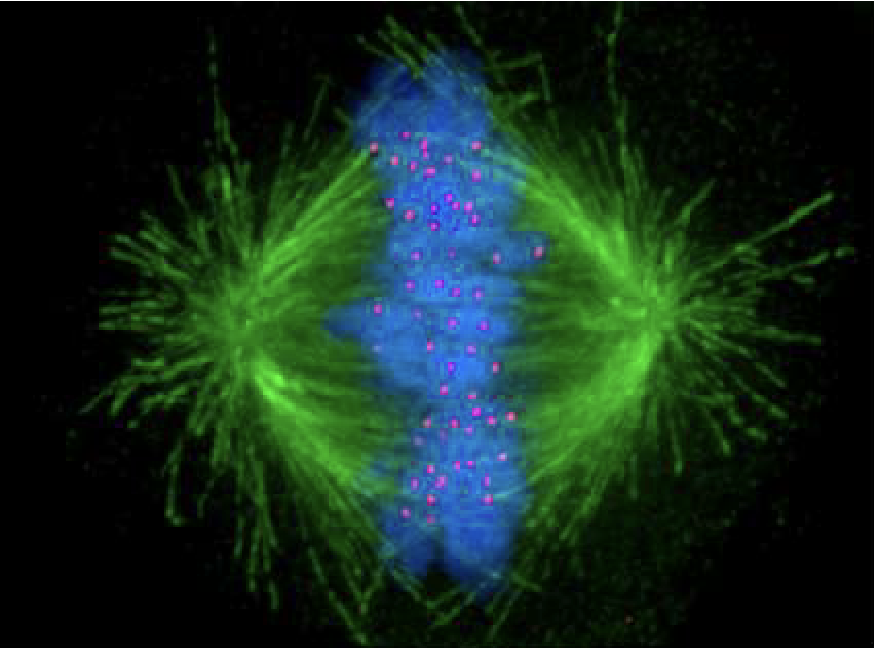
Applied Lecture
Mitotic Spindle Dynamics
What are the key components of the mitotic spindle and their functions?
Metaphase
Green = Microtubules
Blue = DNA
Red = Kinetochores (anchor centromeres to microtubules)
Cells use microtubule dynamics + motor proteins to build spindle
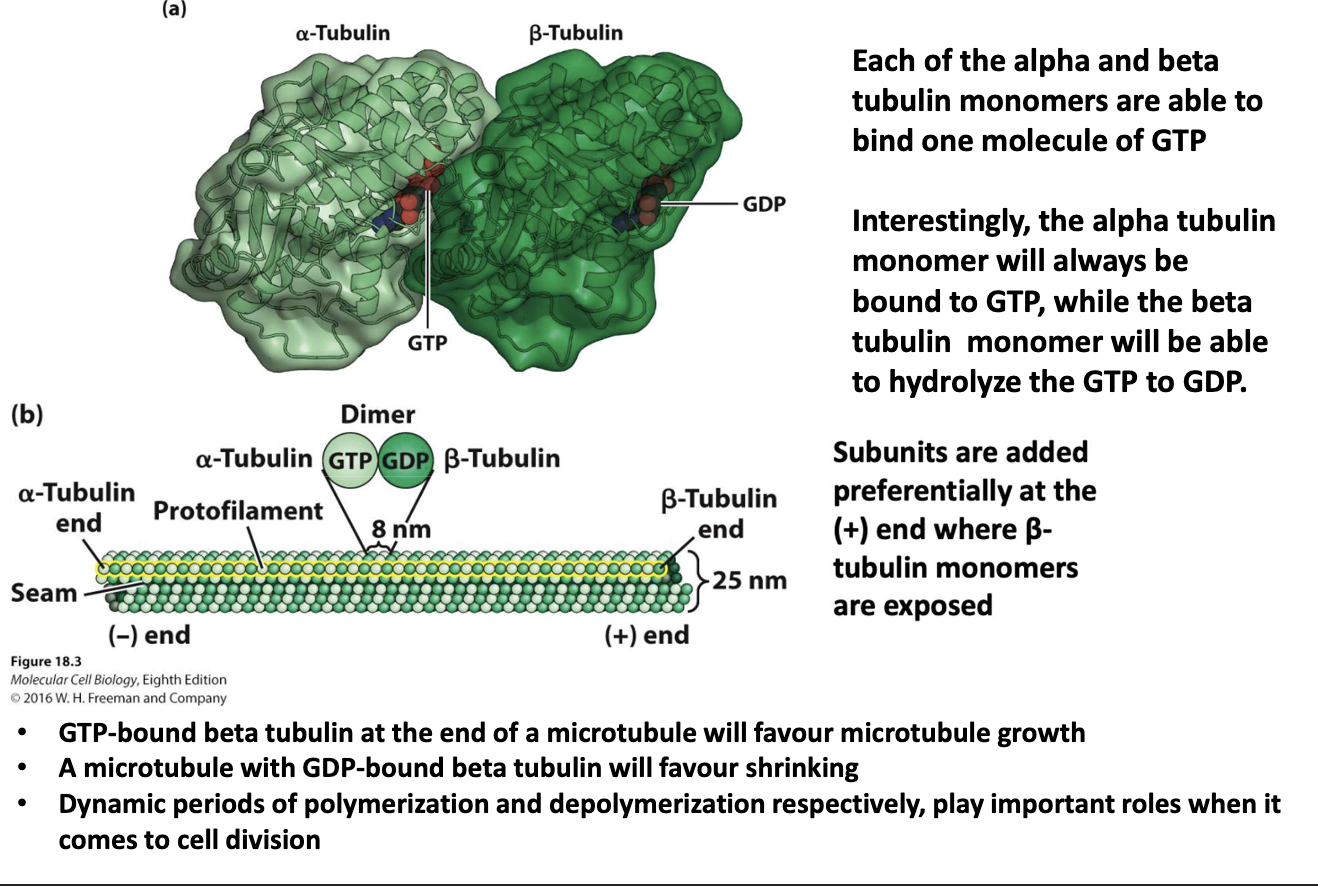
How are microtubules dynamic and polar?
Alpha-tubulin: always bound to GTP (-) end
Beta-tubulin: can hydrolyze GTP → GDP (+) end
GTP-β-tubulin → promotes growth
GDP-β-tubulin → promotes shrinkage
Subunits added at (+) end (β-tubulin exposed)
Growth/shrinkage critical during cell division
What is the basic structure and behavior of microtubules?
Made of 13 protofilaments
Undergo assembly/disassembly
Dynamic behavior varies with cell cycle phase
Both growth & shrinkage occur during mitosis

What is the direction of movement for motor proteins in cell division?
Motor proteins = essential for mitosis
Dynein: moves toward (–) end to cell interior
Kinesin: moves toward (+) end to cell membrane
Both have head domain, coiled coin, and tail domain for cargo
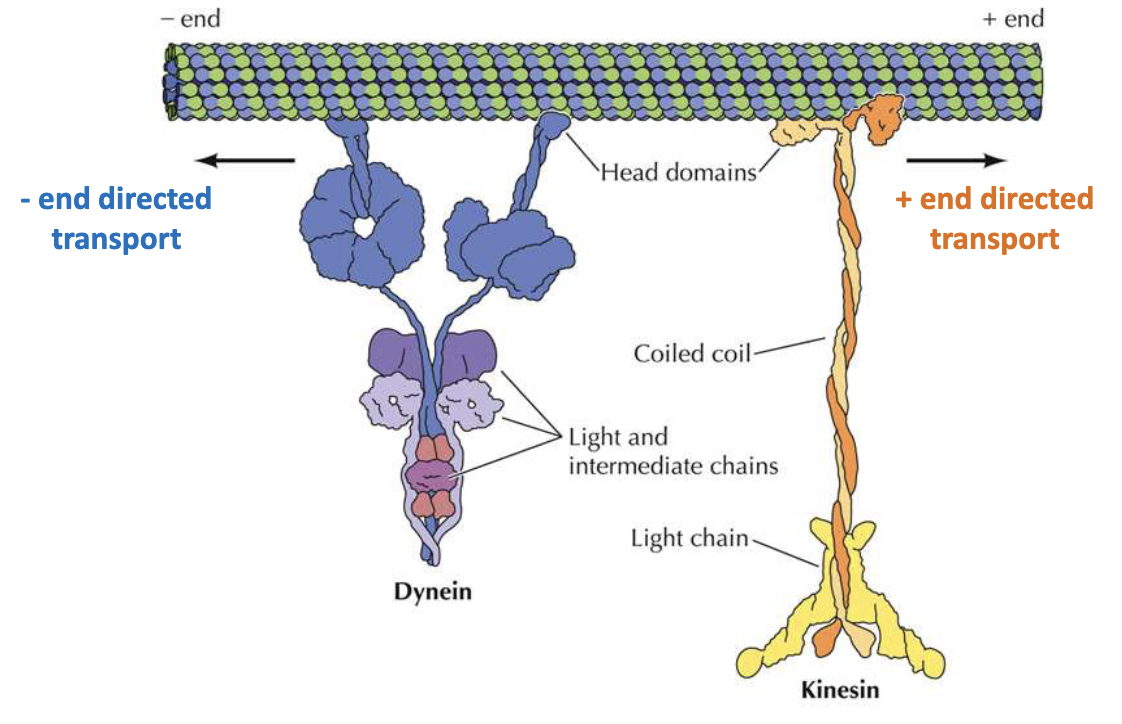
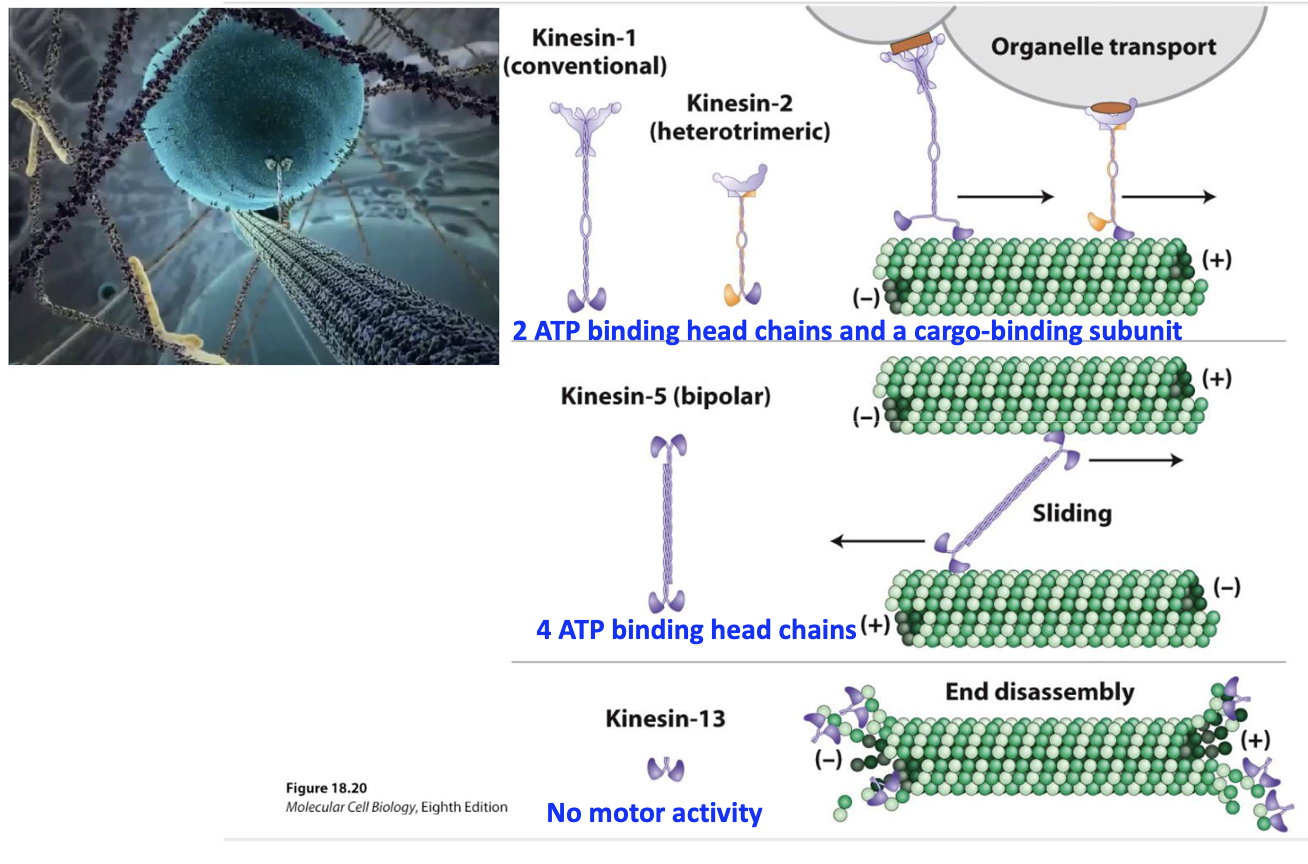
What are the key types and functions of kinesins?
Kinesin-1/2: Organelle transport (2 ATP heads + cargo tail)
Kinesin-5: Bipolar, slides MTs apart (4 ATP heads)
Kinesin-13: No motor activity, promotes end disassembly
Usually seen in (-) end, sometimes seen in (+) end
What do microtubules do during mitosis?
Prophase: form bipolar spindle
Prometaphase/Metaphase: attach chromosomes
Anaphase: separate chromatids
Motor proteins assist in all stages
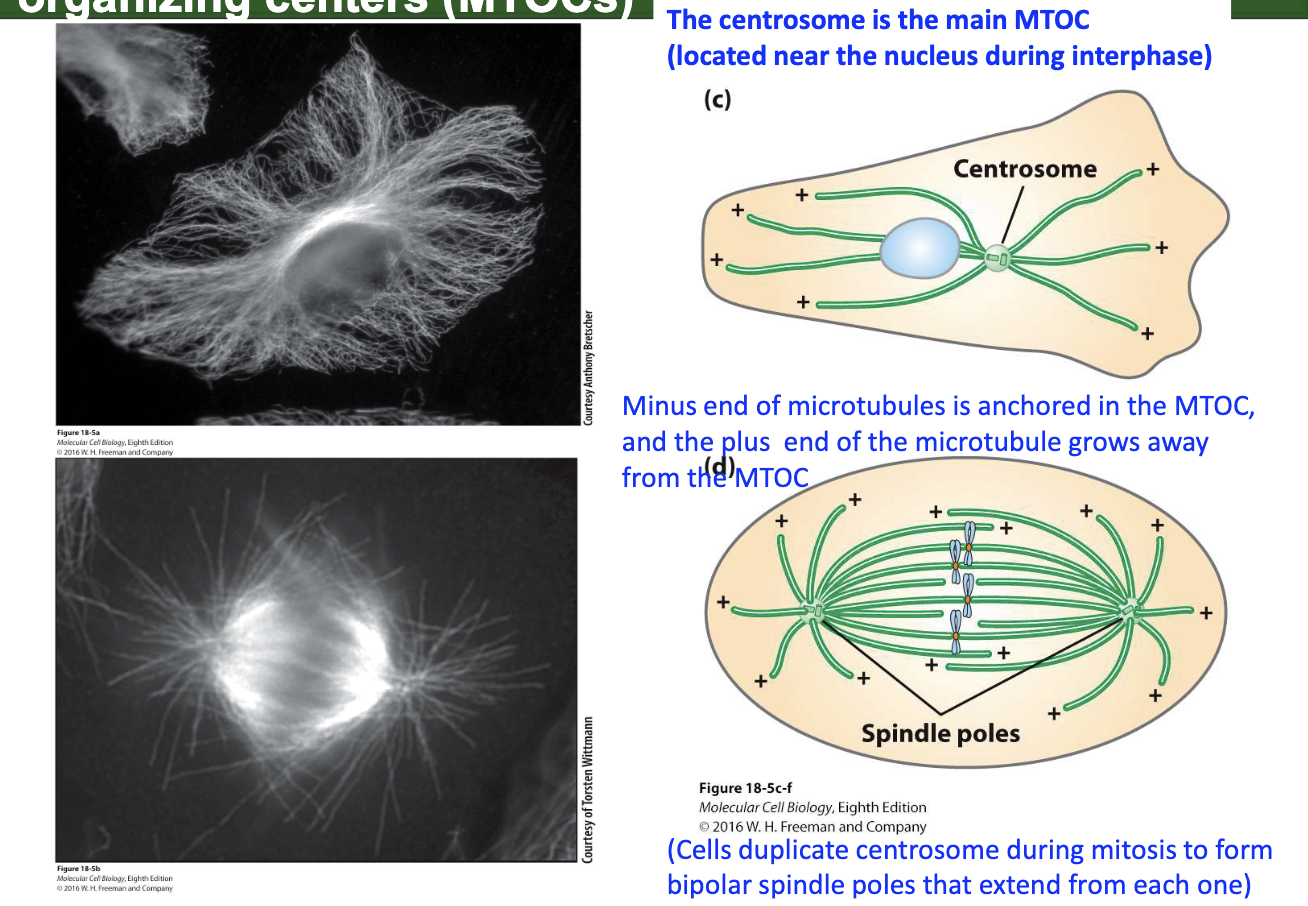
What are MTOCs and their role in spindle formation?
Centrosome = main MTOC (near nucleus in interphase)
MTs grow from + end, anchored at – end in MTOC
Centrosomes duplicate before mitosis → form spindle poles
How do centrosomes help assemble the spindle?
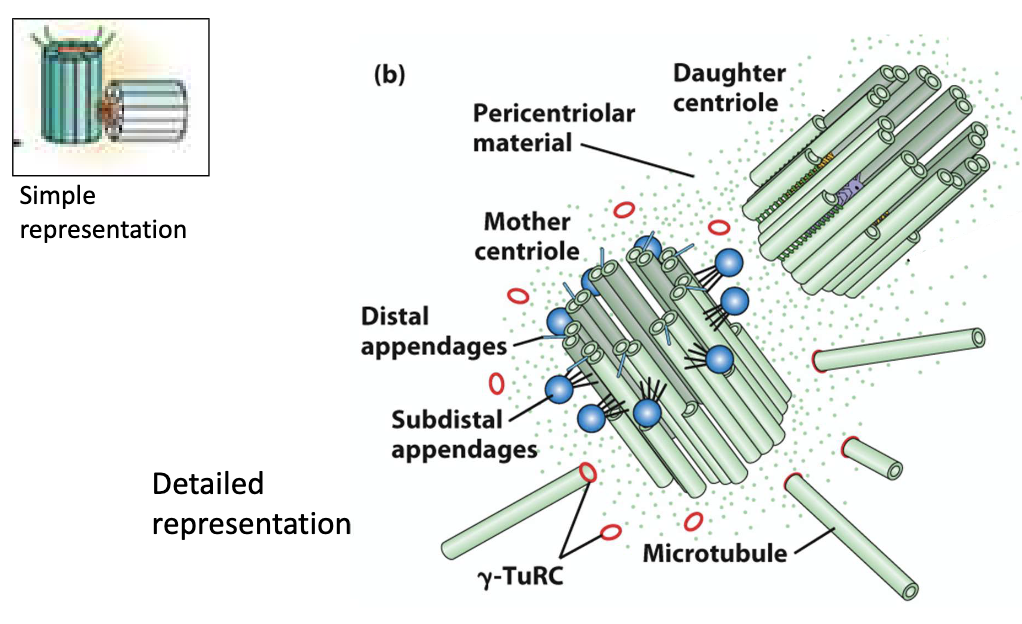
Each centrosome has 2 centrioles (mother and daughter perpendicular)
Around the centrioles is a cloudy area called the PCM (pericentriolar material) — this is where microtubules (MTs) grow from.
The PCM contains γ-TuRC (gamma-tubulin ring complex).
→ This acts like a "baseplate" that helps start microtubule growth.Microtubules start growing from their minus (–) ends in the PCM, and extend outward from their plus (+) ends, forming the spindle.
When and how do centrosomes duplicate?
Happens during G1/S phase, alongside DNA replication
Triggered by CDKs + Plk4 kinase
Centrioles separate → new daughter centrioles bud
G2 phase: daughter centriole growth completes

How do centrosomes separate during mitosis?
Triggered by M phase CDKs
Each centrosome nucleates MTs → becomes a spindle pole
Centrioles move to opposite sides
Occurs in prophase, before nuclear envelope breakdown

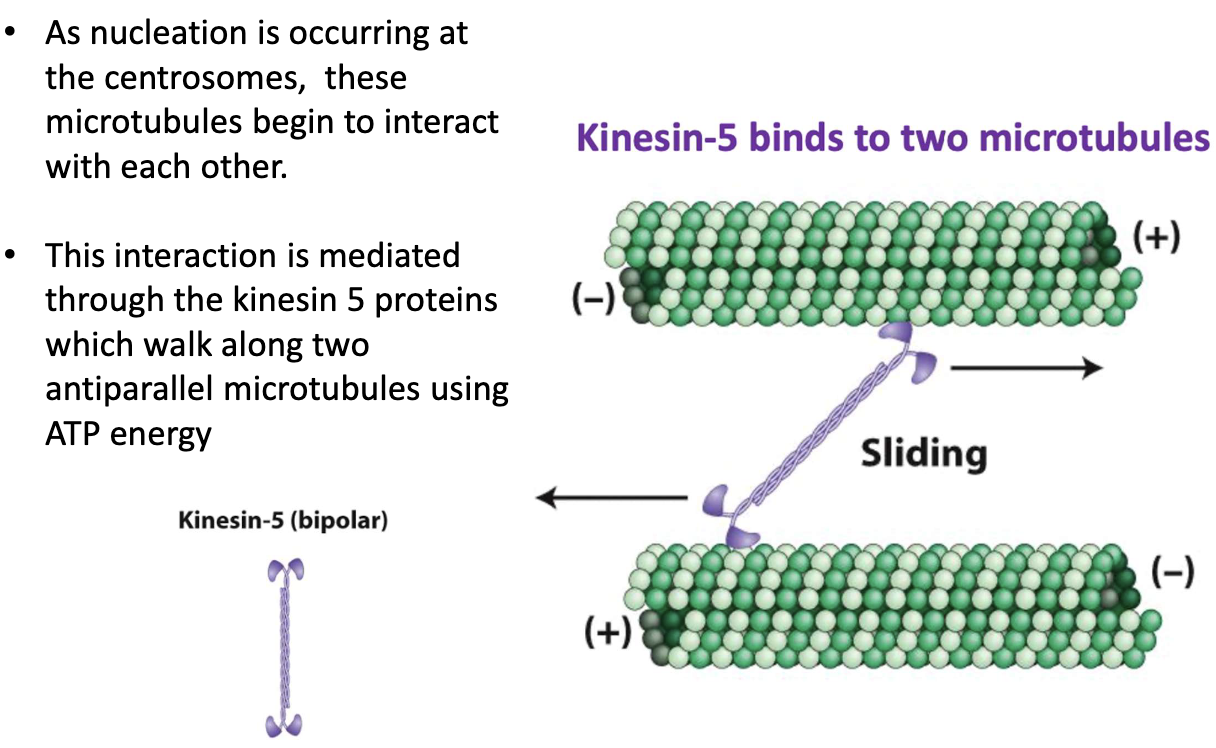
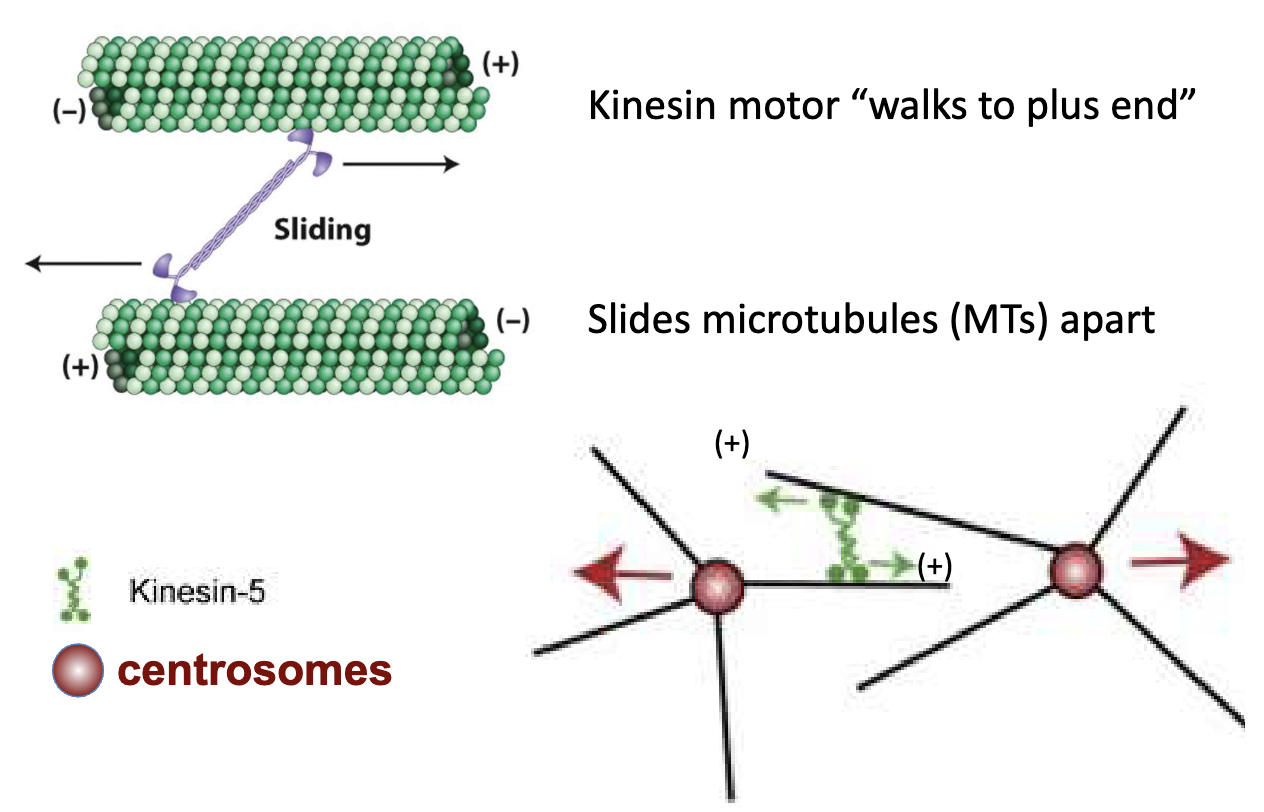
How does kinesin-5 help separate centrosomes?
Microtubules nucleate at centrosomes
Kinesin-5 binds 2 antiparallel MTs
Uses ATP to "walk" toward + ends
Sliding action pushes centrosomes apart
What is the model for centrosome separation?
Kinesin-5 walks toward + ends of MTs
Slides overlapping MTs apart
Centrosomes are pushed to opposite poles
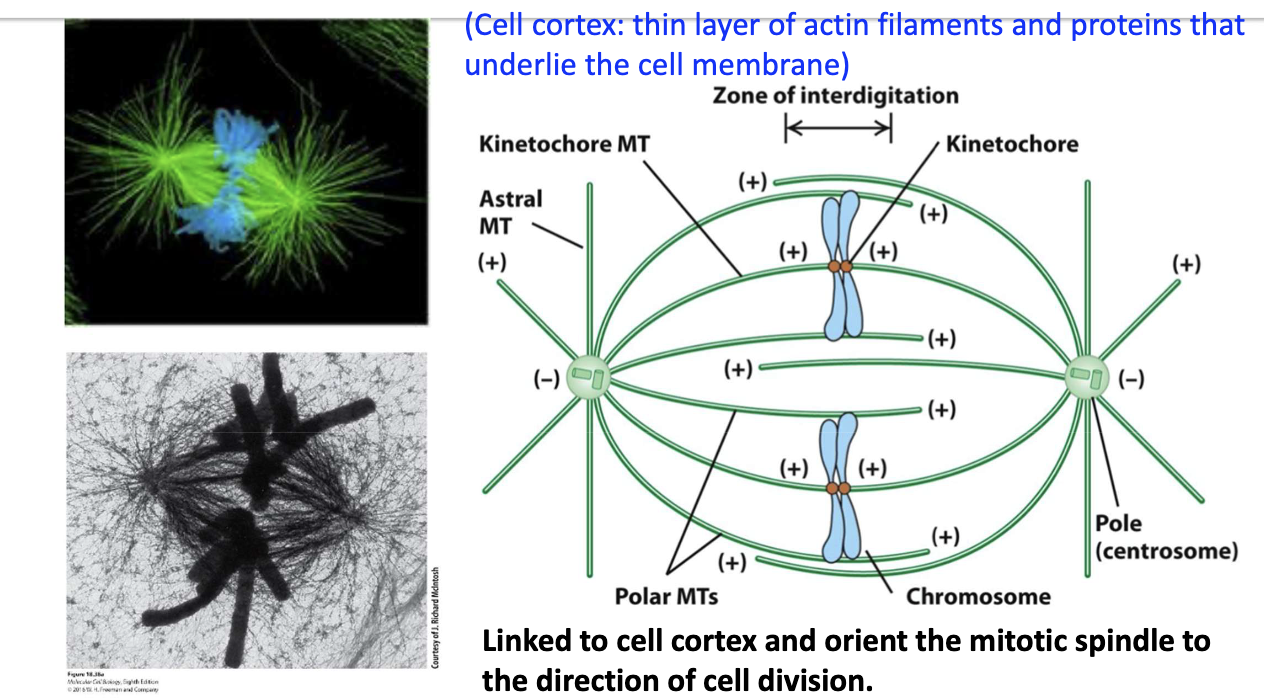
What are the 3 types of spindle MTs and their roles?
Astral MTs – link spindle to cell cortex
Kinetochore MTs – attach to chromosomes
Polar MTs – overlap in center; push poles apart (anaphase)
Different locations, same structure.
Cell cortex: thin layer of actin filaments and proteins that underlie the cell membrane
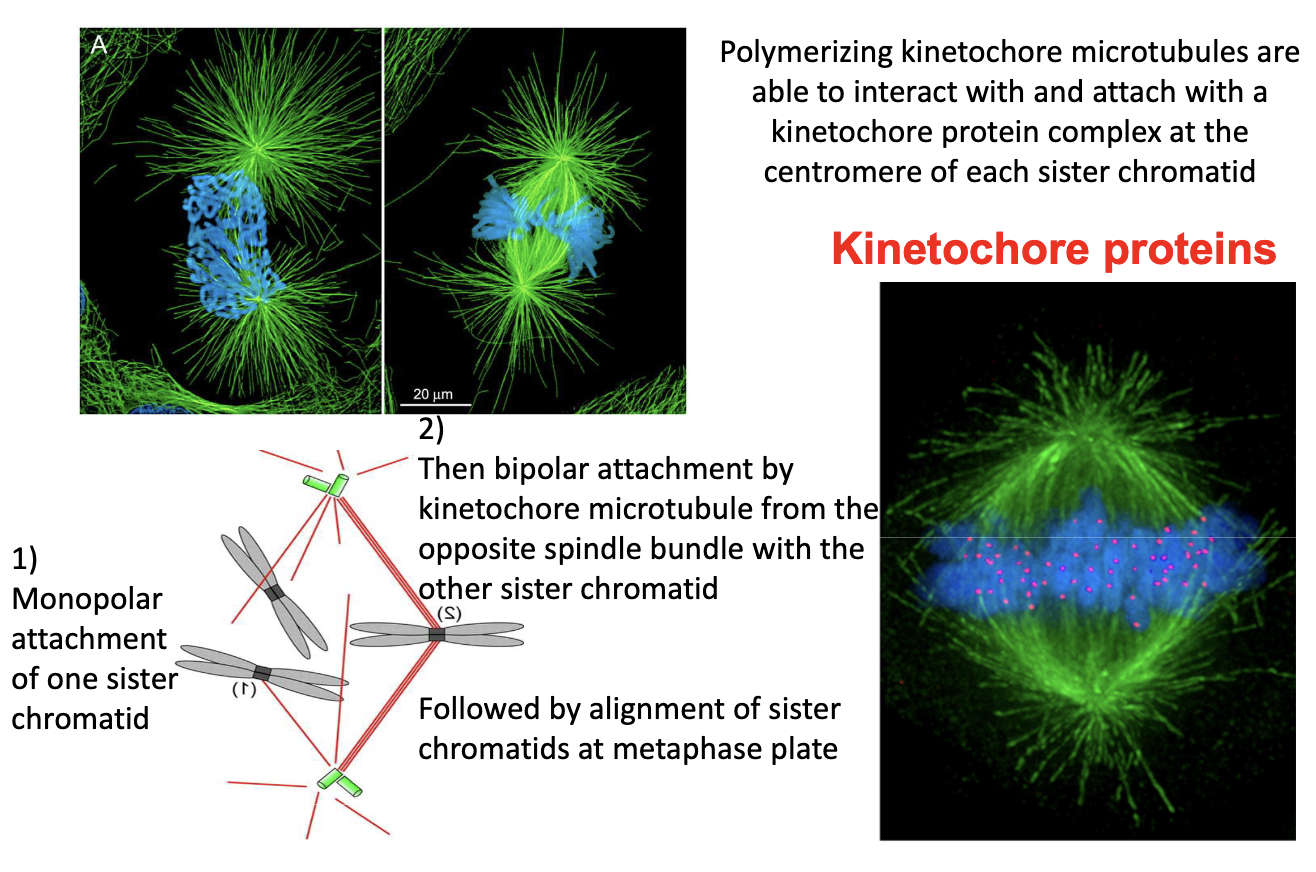
How do kinetochore MTs capture chromosomes?
Prometaphase: MTs attach to kinetochores at centromeres
First: monopolar attachment
Then: bipolar attachment → metaphase plate alignment
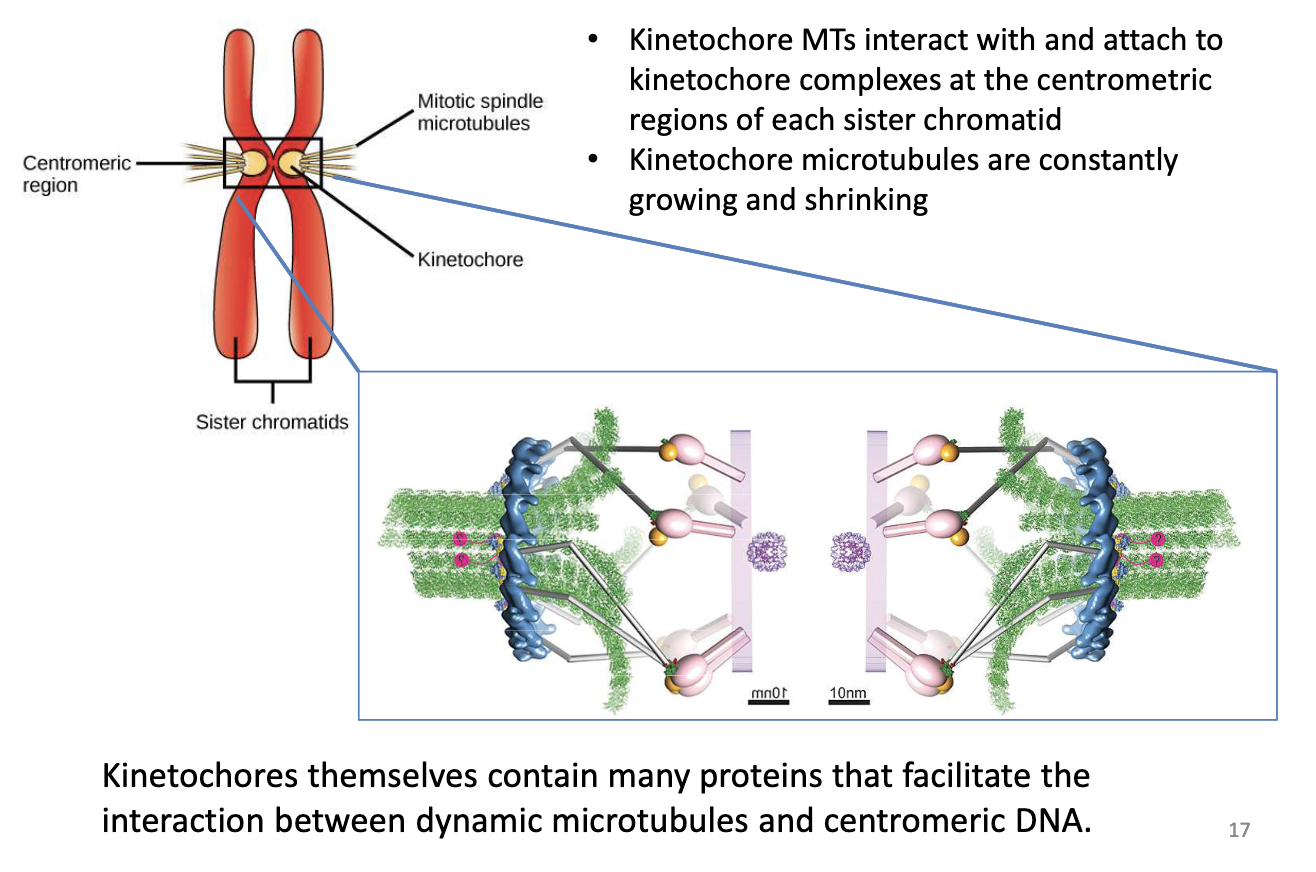
What is the role of kinetochore MTs in mitosis?
Attach to kinetochore protein complexes at centromeric regions of each sister chromatid
Undergo dynamic growth/shrinkage
Kinetochores contain proteins to stabilize MT-chromosome connection
Dynein motor proteins connected to kinetochore complex + sister chromatid, walk towards (-) end spindle poles
Kinesin 13 depolymerizing
How is bipolar attachment achieved?
2 Stages:
Kinetochore MT binds one sister chromatid
Dynein moves sister chromatid pair to allow opposite MT to bind second chromatid
Spindle assembly checkpoint monitors attachment + tension
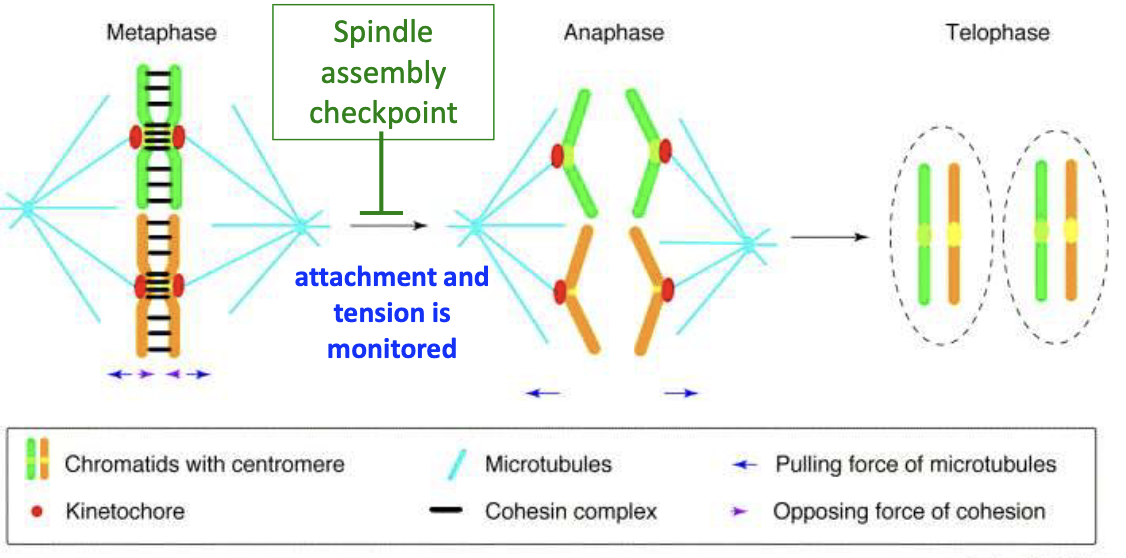
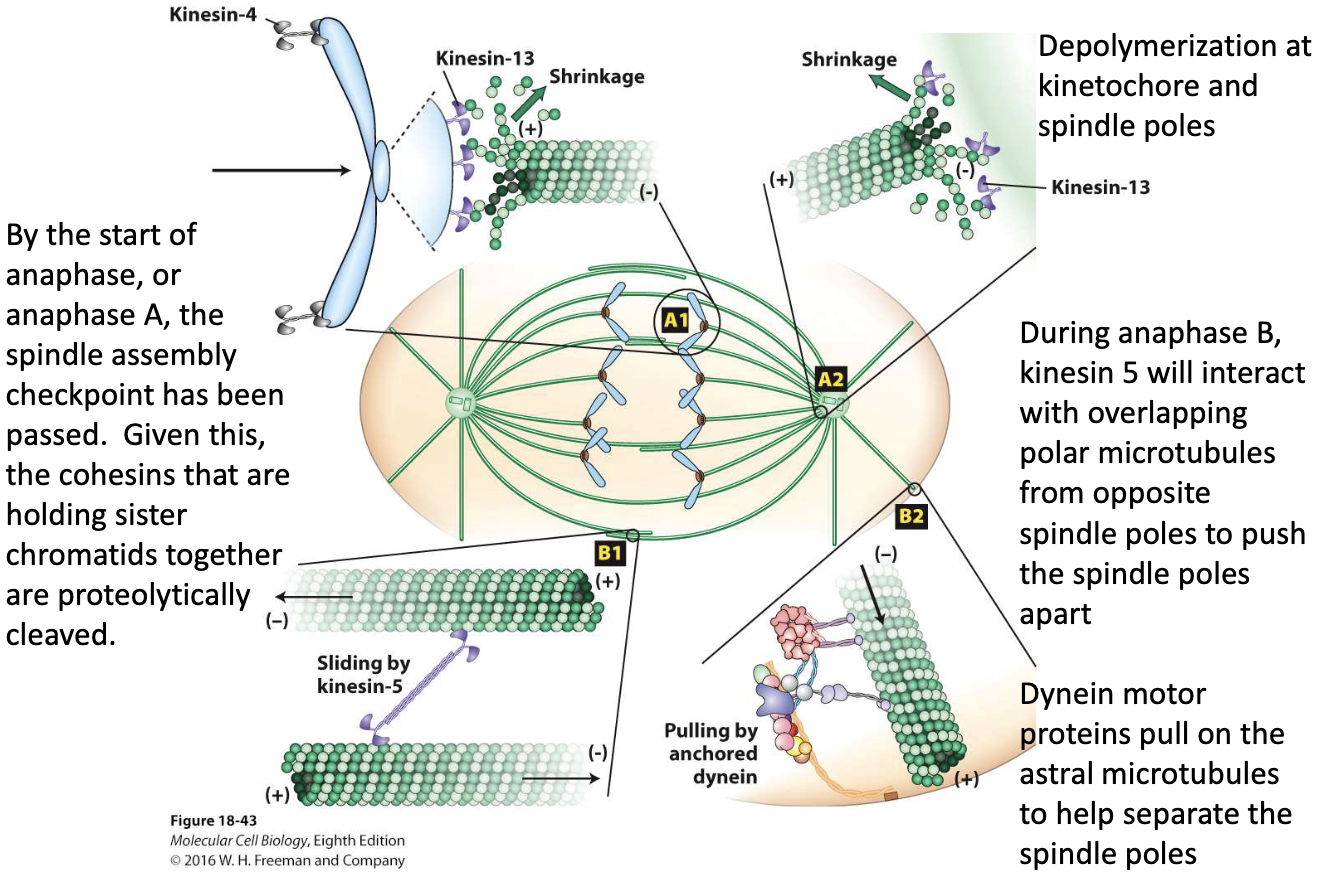
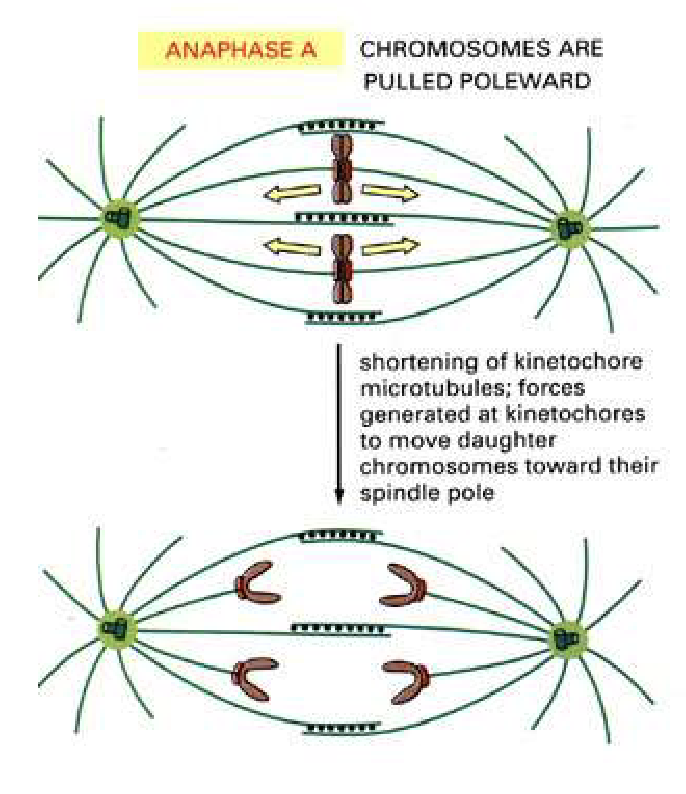
What happens during Anaphase A?
Kinetochore MTs shorten
Kinesin 13 depolymerizing
Forces at kinetochores pull chromosomes to poles
Daughter chromosomes move poleward
What happens during Anaphase B?
Spindle poles move apart
2 forces involved:
Kinesin-5 slides overlapping polar MTs apart
Dynein pulls on astral MTs at cortex
Polar MTs grow at + ends

What key events occur during anaphase?
Anaphase A:
Cohesins cleaved
Chromosomes pulled to poles (MT depolymerization at kinetochores by Kinesin-13)
Anaphase B:
Kinesin-5 slides polar MTs
Dynein pulls astral MTs
Helps move the two spindle poles farther apart stretching the spindle and separating the chromosomes.