BIOL365- Weeks 1-7
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
14 Terms
5 Kinds of genetic mutations
Single nucleotide mutations, Simple insertions or deletions, Duplications & large mutations, Tandem repeat mutations, Transposable elements
Single nucleotide mutations
a change in a single nucleotide (A, T, C, or G) in a DNA sequence
Simple insertions or deletions
Nucleotides added or removed from a sequence
ie. deleting G or adding in an A into the existing DNA sequence
Tandem repeat mutations
a sequence of two or more DNA bases that is repeated numerous times in a head-to-tail manner on a chromosome
ie. ACAGCAGCAGT
Duplications & large mutations
significant sources of genetic variation, often involving the copying or rearrangement of large DNA segments. These mutations can lead to the creation of new genes, alter gene expression, and impact overall genome structure
Transposable elements
"jumping genes," are DNA sequences that can move from one location to another within a genome
Sources of genetic variation for a population
migration and mutation. The latter is the ultimate source
Heterozygosity
the state of having two different versions (alleles) of a gene at a particular locus on a chromosome, one inherited from each paren
Hardy-Weinberg Principle 5 assumptions
No Mutation: New alleles are not introduced into the population.
No Gene Flow: There is no migration of individuals into or out of the population.
Random Mating: Individuals mate randomly, without any preference for certain genotypes.
Large Population Size: The population is large enough to prevent random fluctuations in allele frequencies (genetic drift).
No Natural Selection: All genotypes have equal survival and reproductive rates.
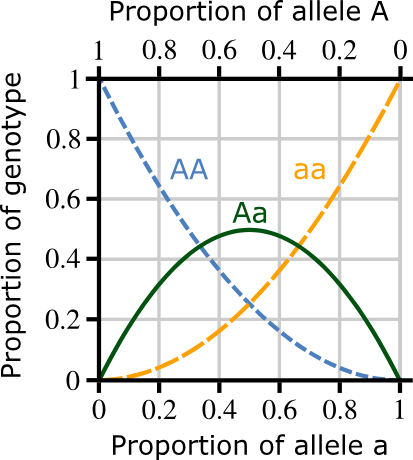
Hardy-Weinberg Principle calculations
p + q = 1, where 'p' is the frequency of one dominant allele and 'q' is the frequency of the other recessive allele.
p² + 2pq + q² = 1, where p² is the frequency of the homozygous dominant genotype, 2pq is the frequency of the heterozygous genotype, and q² is the frequency of the homozygous recessive genotype
Standard neutral theory
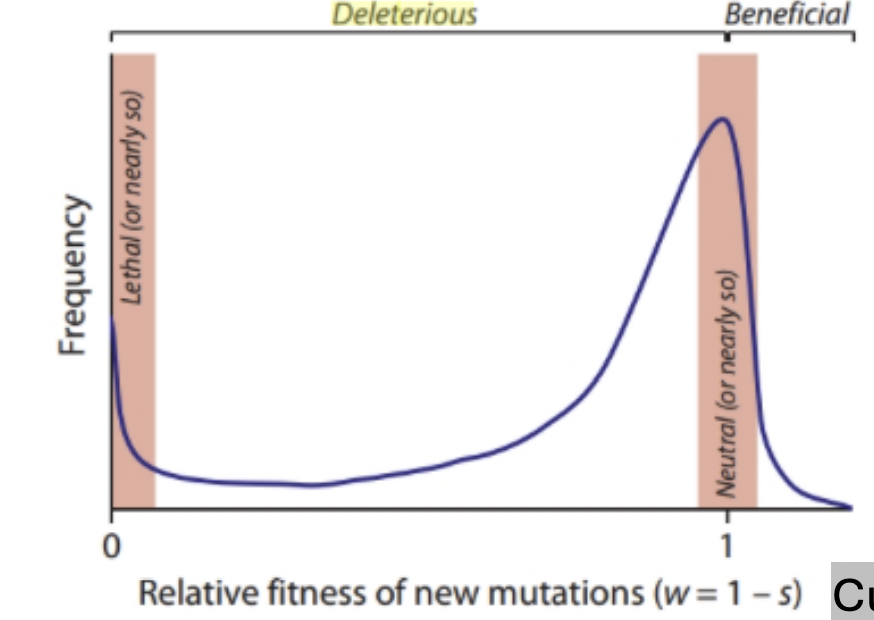
most evolutionary changes at the molecular level are driven by random genetic drift of neutral or nearly neutral mutations, rather than positive selection acting on advantageous traits
Relative fitness of new mutations
The molecular clock
to estimate the time of divergence between different species or lineages based on the rate of molecular evolution, primarily through analyzing DNA or protein sequences
for example: in 2 species The sequence is 1,000 base pairs (bp) long., After alignment, you find 50 nucleotide differences between the two sequences, You know from previous fossil or genetic calibration studies that the mutation rate for this segment is approximately 1 × 10⁻⁸ substitutions per site per year. You assume mutations have occurred independently in both lineages, so the divergence rate is 2 × mutation rate.
= The two species diverged approximately 2.5 million years ago.

Nearly neutral theory
most molecular evolution is driven by genetic drift acting on slightly deleterious mutations, rather than being purely neutral or driven by strong selection.
It bridges the gap between the neutral theory, which posits that most mutations are neutral, and the selection theory, which emphasises the role of strong selection