exam 2 flashcards
1/85
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
86 Terms
When does DNA replication happen?
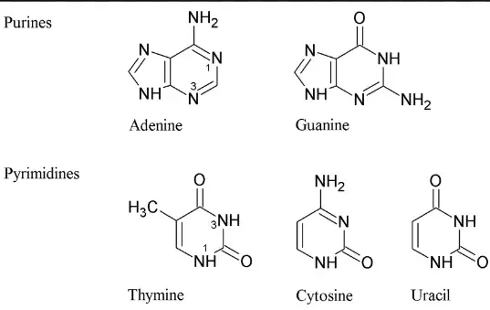
Purines
One of the two categories of nitrogenous bases in nucleic acids, including adenine and guanine.
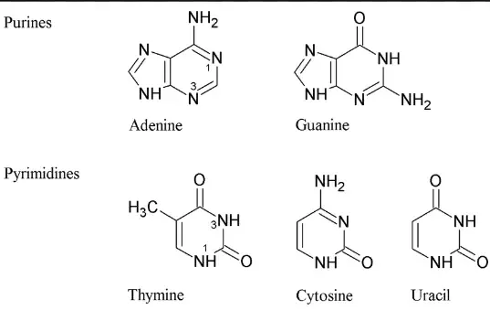
Pyrimidines
One of the two categories of nitrogenous bases in nucleic acids, including cytosine, thymine, and uracil.
Transformation
A process by which genetic material is transferred between bacteria, leading to changes in their genotype or phenotype.
“Zombies”

Frederick Griffith Experiment (1928)
Showed there was a transforming factor
Tested pathogenic and non-pathogenic strains of pneumonia

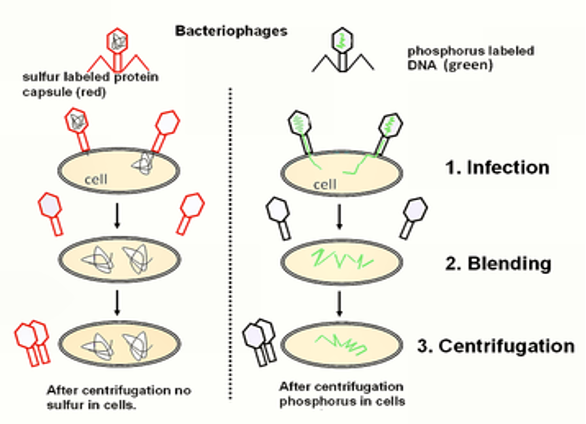
Hershey and Chase experiment (1944)
Showed DNA and not protein is genetic material
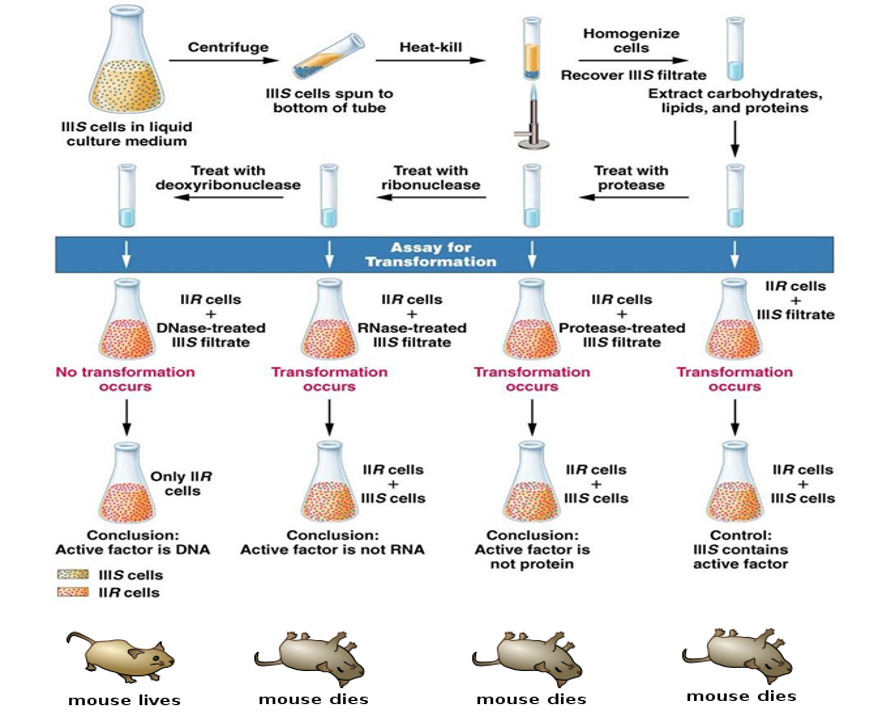
Avery experiment (1944)
Provided evidence that DNA was the transformation factor
The structure of DNA discovery (1953)
Watson and Crick, and Rosalind Franklin (photo 51)
Identified the double helix structure of DNA, which is essential for genetic information storage and replication.

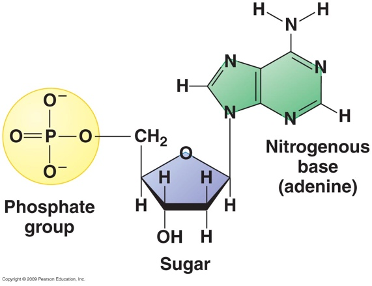
Nucleotides
The building blocks of DNA and RNA, consisting of:
a phosphate group
a pentose sugar
a nitrogenous base
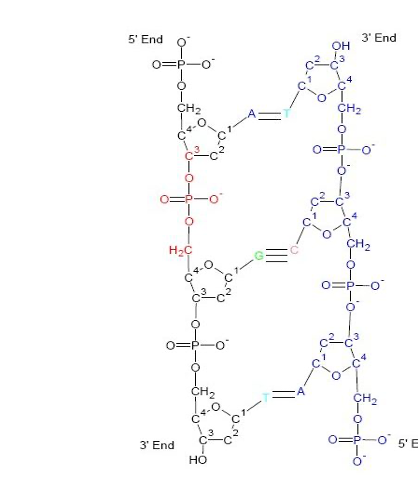
5’ to 3’
A-T linked by two bonds
G-C linked by three bonds
Semiconservative replication
50% of DNA old, 50% new
Parental and daughter strand
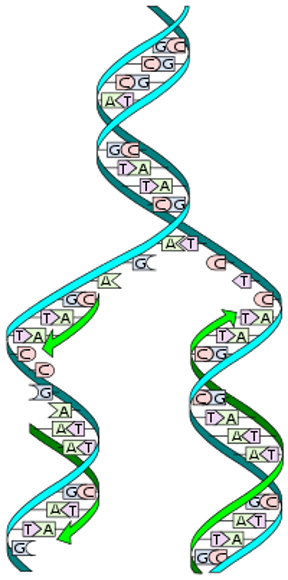
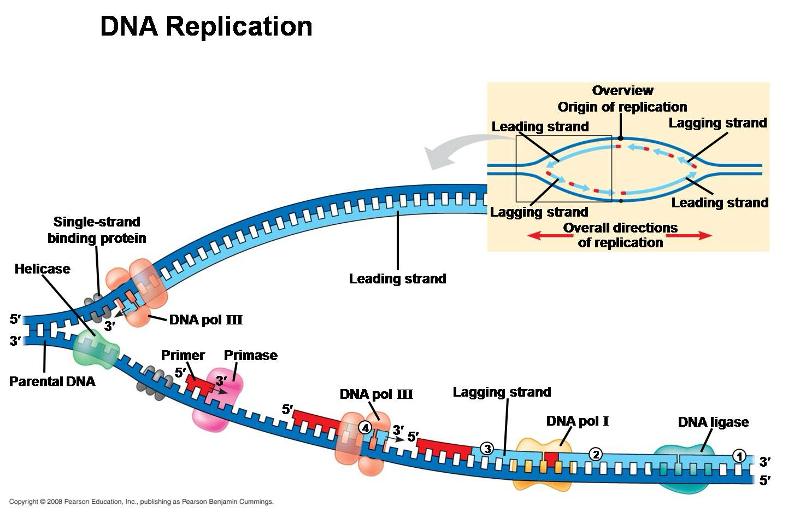
DNA Replication
DNA helix splits into leading strand and lagging strand
Strands grow from 5’ to 3’
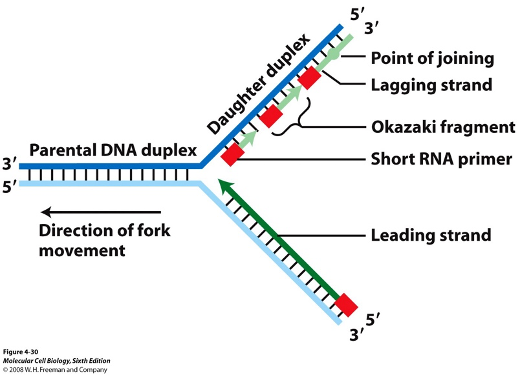
Replication Enzymes
proteins that facilitate the process of DNA replication, including:
helicase
single stranded binding protein
DNA polymerase
primase
ligase
topoisomerase
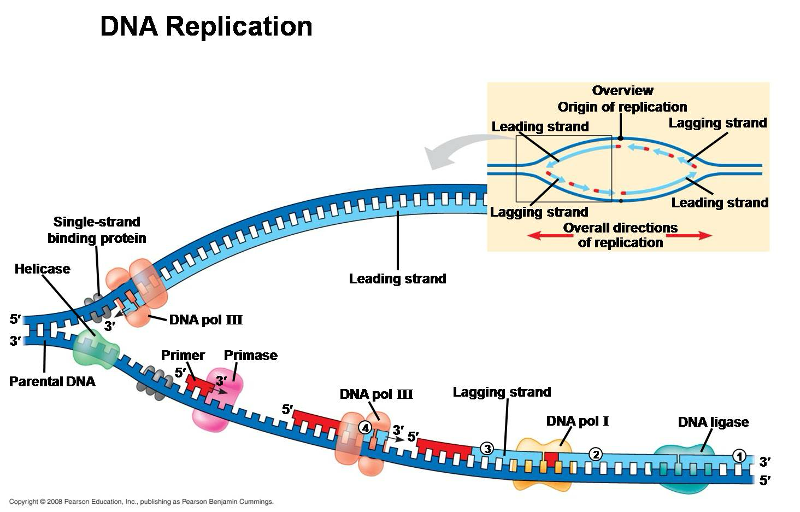
Helicase
Unwinds helix
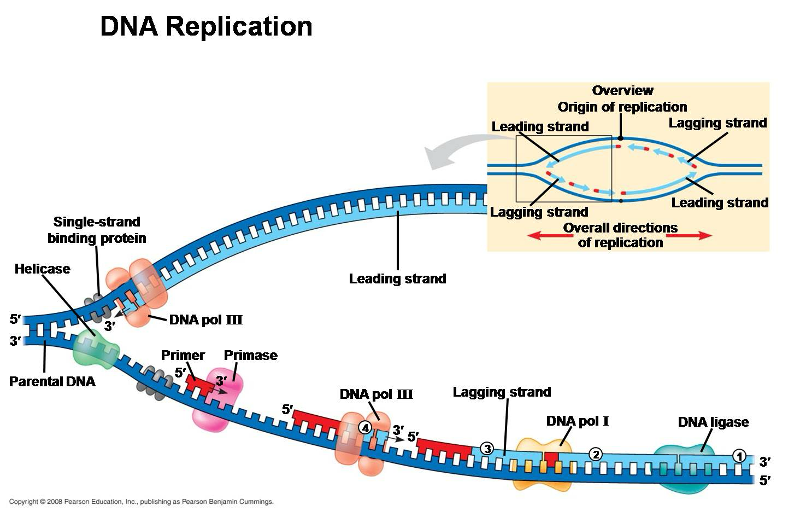
Single stranded binding protein
Prevents helix from coming back together
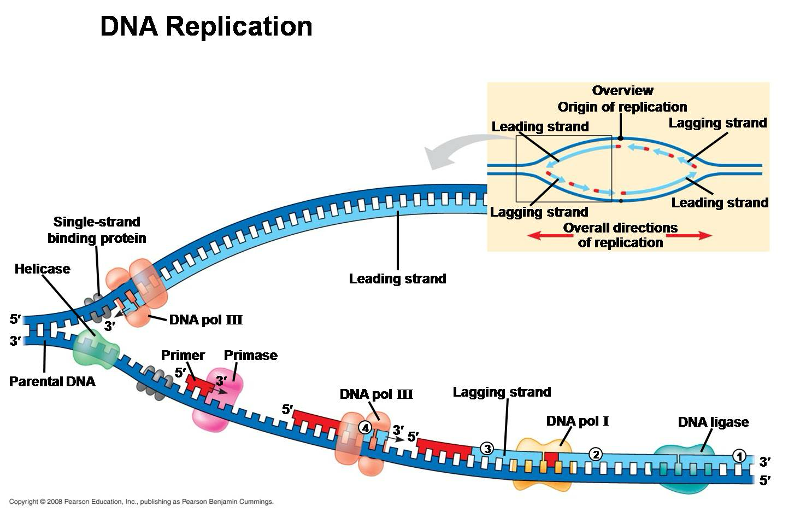
DNA polymerase
Recruits nucleotides
Primase
Guides DNA polymerase
Ligase
Binds Okazaki fragments
Topoisomerase
Prevents supercoiling
When does central dogma / transcription happen?
Anytime
An organism or cell that has a nucleus and membrane-bound organelles.
Organisms that include animals, plants, fungi, and protist.
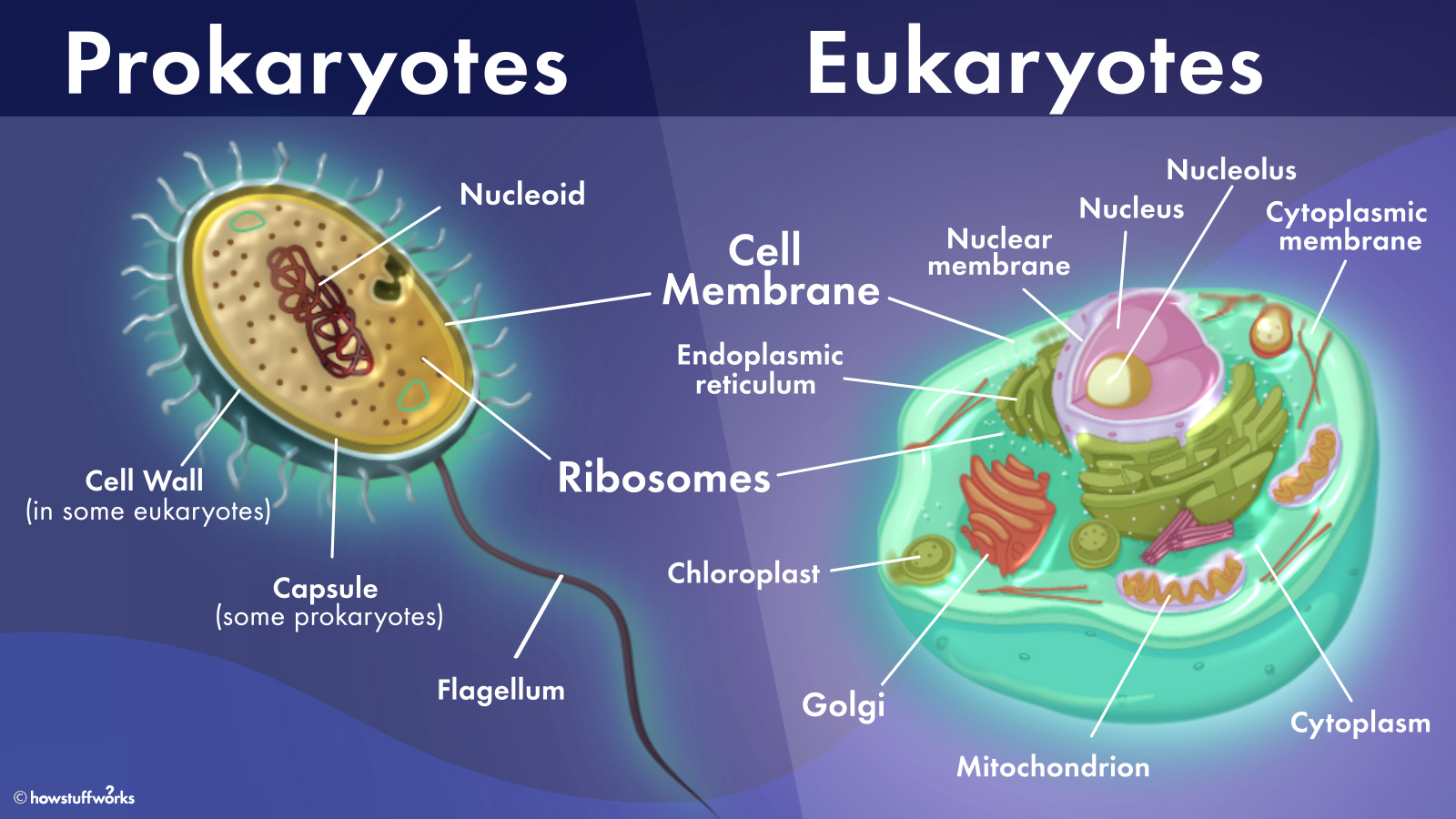
How many cells do we start out as and end at?
We start off as one cell; a zygote; after the sperm fertilizes the egg
~40 trillion cells are in a fully mature adult
Numerous cells must divide and differentiate to create a fully functioning organism
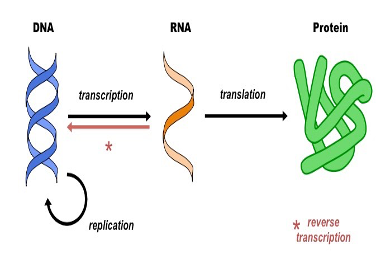
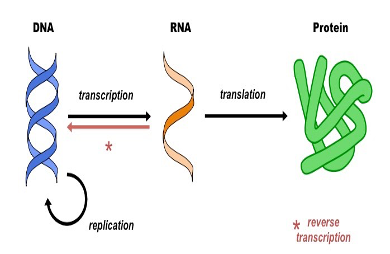
Reverse transcription
RNA to DNA
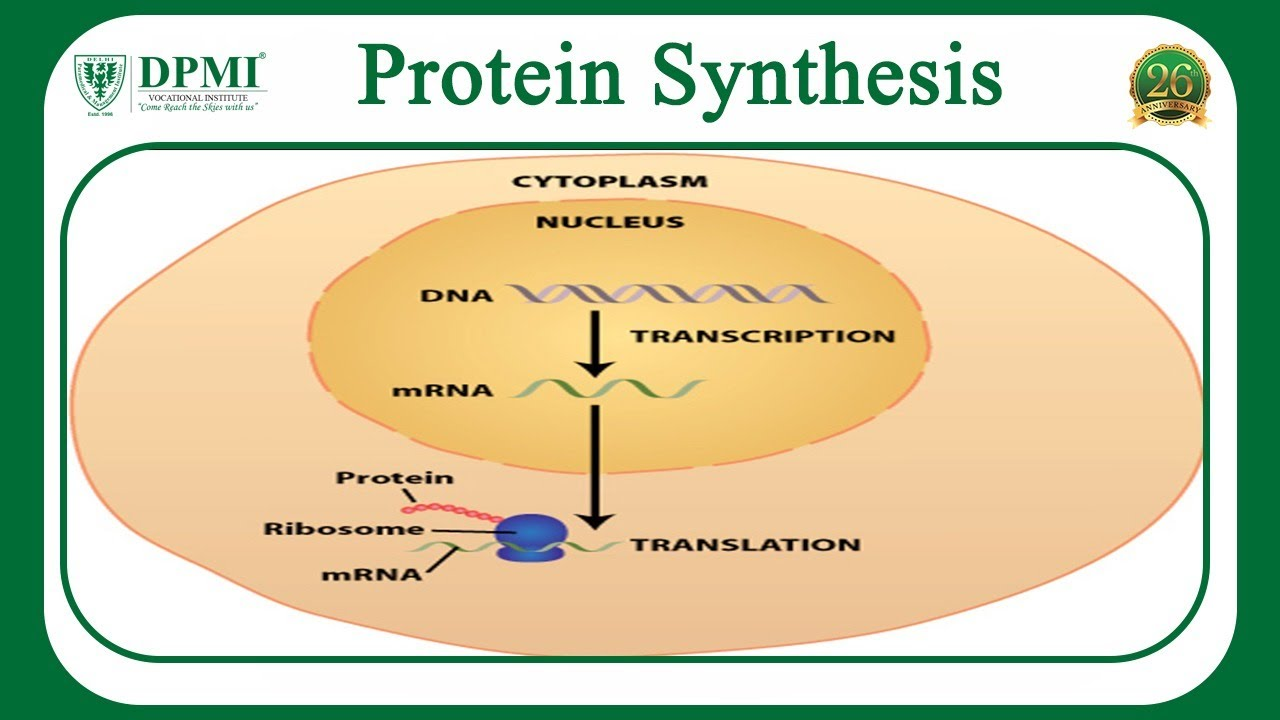
Protein synthesis takes place where?
In the cytoplasm and ribosomes of the cell
(Protein synthesis is the process by which cells create proteins, the building blocks of all living organisms. It involves two main steps: transcription and translation)
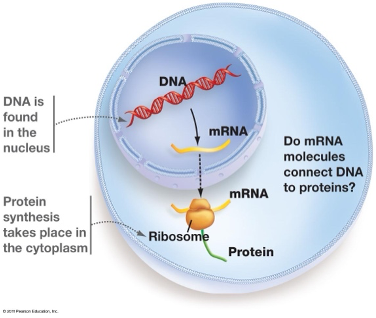
Messenger RNA that carries instructions from DNA to the ribosome to make protein.
Ribosomal RNA, a structural component of ribosomes, needed to form peptide bonds.
Required to recruit RNA initiation complex
key component of transcription
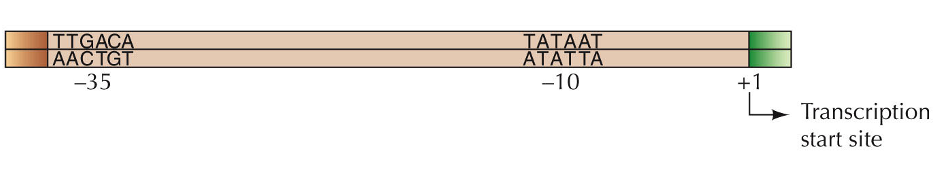
In eukaryotes the primary promoter is the TATA box (consensus sequence).
RNA initiation complex
Needed for RNA polymerase function, one member is a helicase.
TATA binding protein required to bind the promoter.
a key component of transcription
Enzyme that recruits nucleotides to make RNA.

What is the primary promoter in eukaryotes?
The TATA box (consensus sequence)
Promoter: DNA sequence to which RNA polymerase binds (initiation complex binds first). The region upstream of the transcription initiation site has 2 different consensus 6 base pair sequences. One at –10 and one at –35.
Transcription overview sequence
RNA initiation complex bind TATA box.
Specifically the first is TFIID (which contains TATA binding protein (TBP)).
Then RNA polymerase binds to complex.
Lastly remaining members of complex bind,
One member being a helicase.
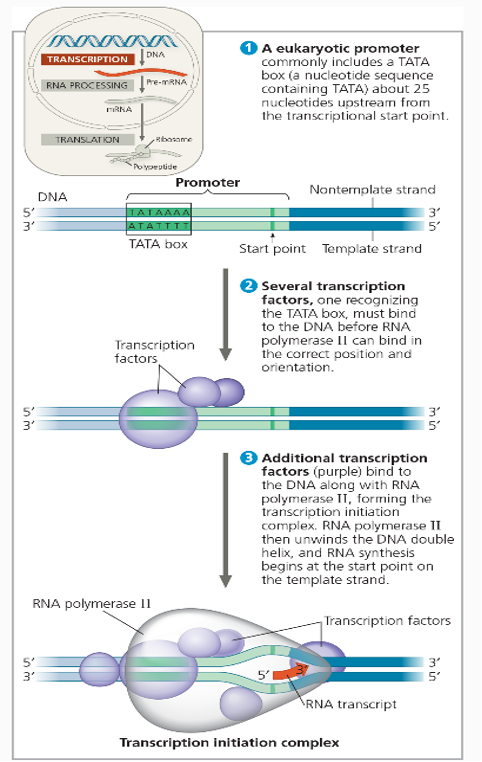
Parts of Transcription sequence
Initiation - After all members of complex are present RNA polymerase starts recruiting nucleotides
Elongation - RNA polymerase moves 5’ to 3’ recruiting nucleotides.
Termination - RNA polymerase reaches termination sequence and then detaches.
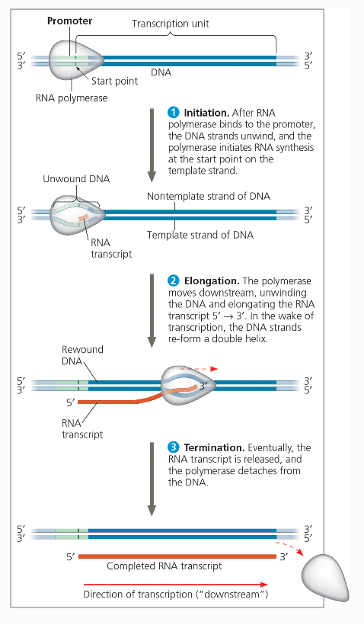
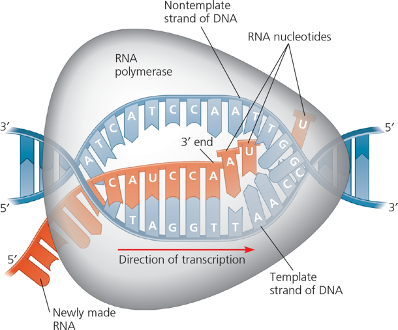
Elongation
RNA polymerase recruit's uracil instead of thymine. RNA polymerase lacks proofreading activity.
How can RNA polymerase afford to lack proofreading activity?
Because RNA is typically synthesized quickly and is often not as critical as DNA, allowing for some errors without significant consequences.
RNA processing and turnover
Transcriptional regulation is the major method to regulate gene expression, but RNA processing and turnover are also important
Pre RNA’s are modified before transport to the cytoplasm. This includes modification of both ends and removal of introns
7-methylguanosine cap
Polyadenylation of tail
Splicing: introns are removed by splicing from exons
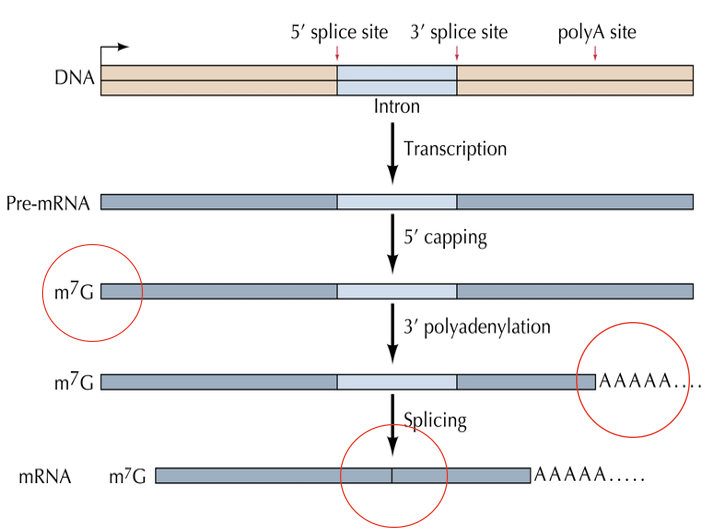
7-methylguanosine cap
a cap is placed on the 5’ end co-transcriptionally.
It stabilizes the RNA and facilitates binding of RNA to the ribosome
Polyadenylation of tail
Polyadenylation is a crucial post-transcriptional modification in eukaryotes where a poly(A) tail;
a sequence of adenine nucleotides, is added to the 3' end of mRNA, enhancing its stability, facilitating export from the nucleus, and promoting translation
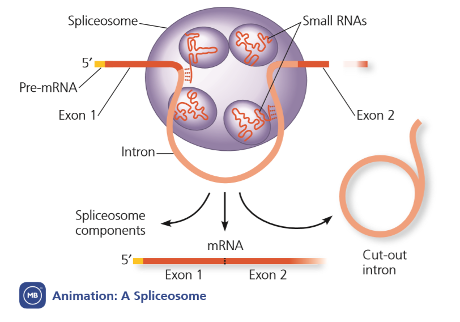
Ribozymes
bind to start of mRNA molecules, which carry the genetic code from DNA to the ribosome.
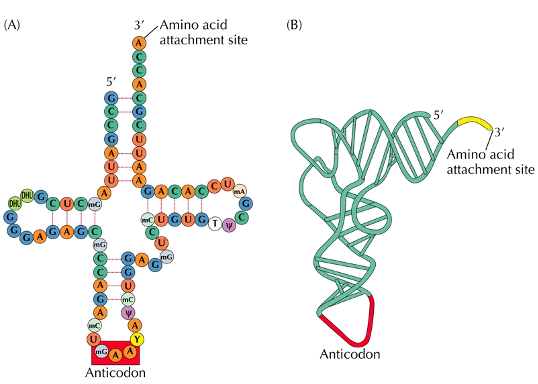
Transfer RNAs (tRNAs)
needed for translation
transfer RNA; a small RNA molecule in protein synthesis that acts as an adaptor molecule, linking mRNA codons to specific amino acids.
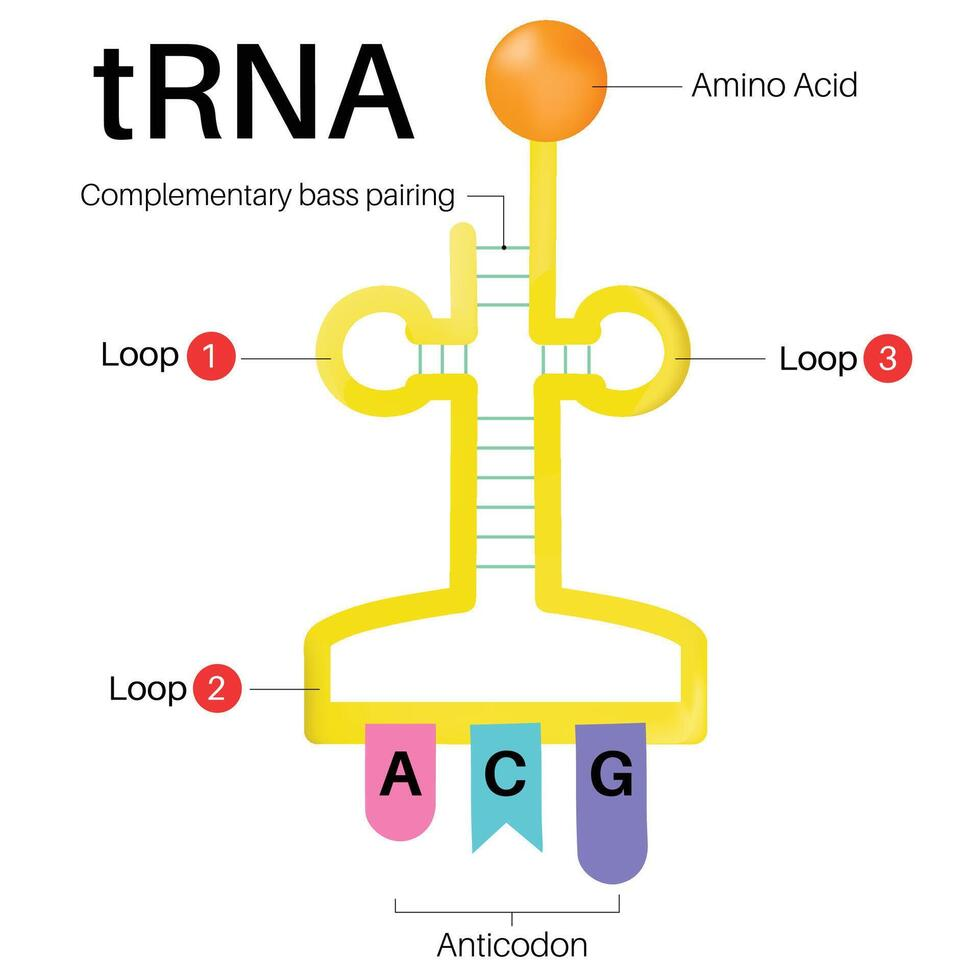
Two major sites in transfer RNA translation
an amino acid attachment site and an anticodon loop located at the opposite end to bind the right codon
Why are ribosomes needed for translation?
To catalyze protein synthesis.
Two subunits composed of protein and RNA.
Abundant (1x106 in eukaryotic cells)
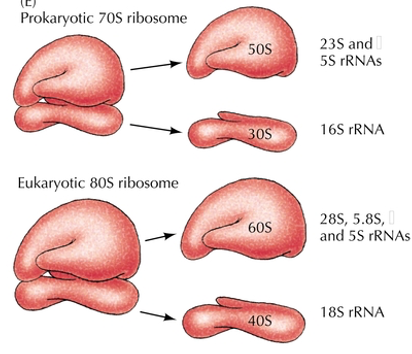
Ribosomal RNA deficiency disease
Treacher Collins Syndrome- Mutated TCO1 gene which is required for rRNA transcription.
Initiation , elongation , and termination
Initiation is complex in eukaryotes (>12 initiation factors) but simpler in prokaryotes.
Starts when tRNA-methionine and mRNA bind to the small ribosomal subunit
The peptide chain elongates by subsequent addition of AAs as the ribosome moves down the mRNA. When a stop codon is reached (UAA), the reaction terminates, the protein is released and the ribosome dissociates from mRNA.
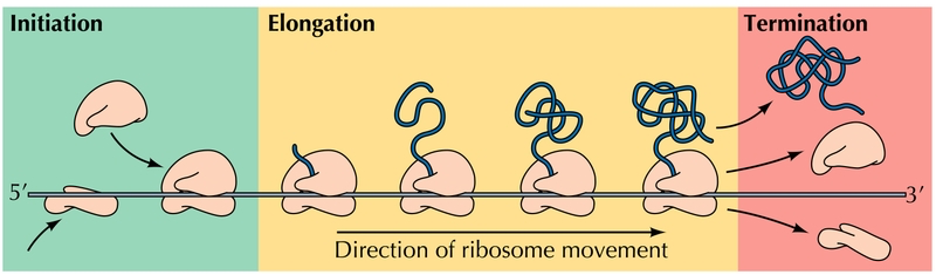
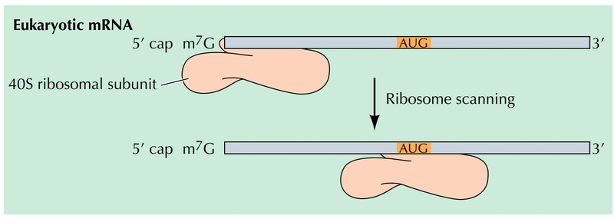
Initiation of translation
Methionine encoded by AUG, initiates translation in both prokaryotes and eukaryotes
The 7- methyl guanosine cap of eukaryotic mRNA binds the ribosome and helps to align it for translation
Initiation, elongation and termination factors
The first step in prokaryotes is binding of 3 initiation factors to the ribosome. The mRNA and tRNA join, GTP is hydrolyzed and the 70S initiation complex is formed
Initiation in eukaryotic cells is complex and requires at least 12 initiation factors. After initiation, elongation and termination factors are required for translation to be completed
Elongation of the peptide chain
Start with first amino acid, binding site, found in bacteria,
first amino acid comes to the P site
Everything else comes to the A
Peptide bond forms through dehydration reaction
Entire ribosome subunit shifts which pushed amino acid
First amino acid pops off and are then in the chain?
To create long polypeptide chain
Continues until release factor binds to ?
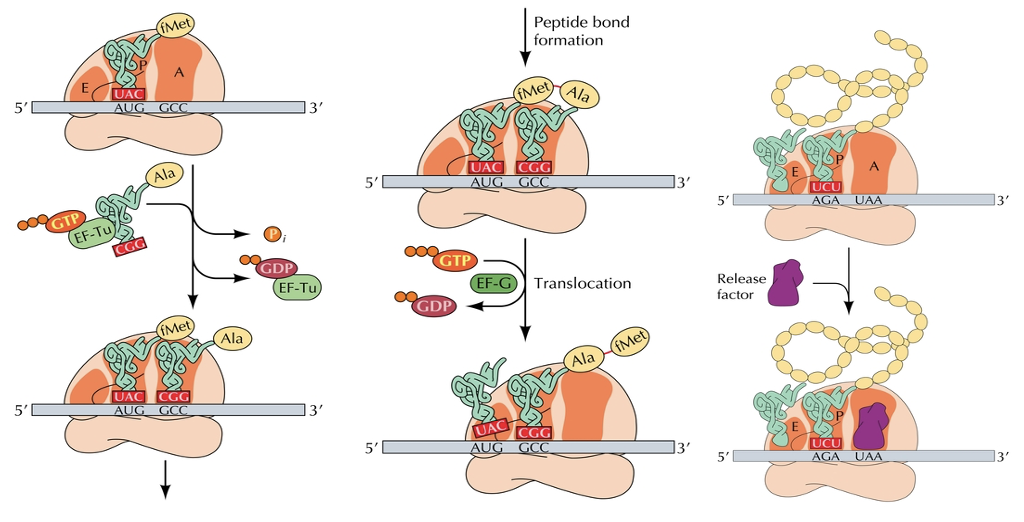
Termination of translation
The release factor breaks the bond between the peptide chain and the tRNA
The nascent protein, tRNA, and ribosome all dissociate
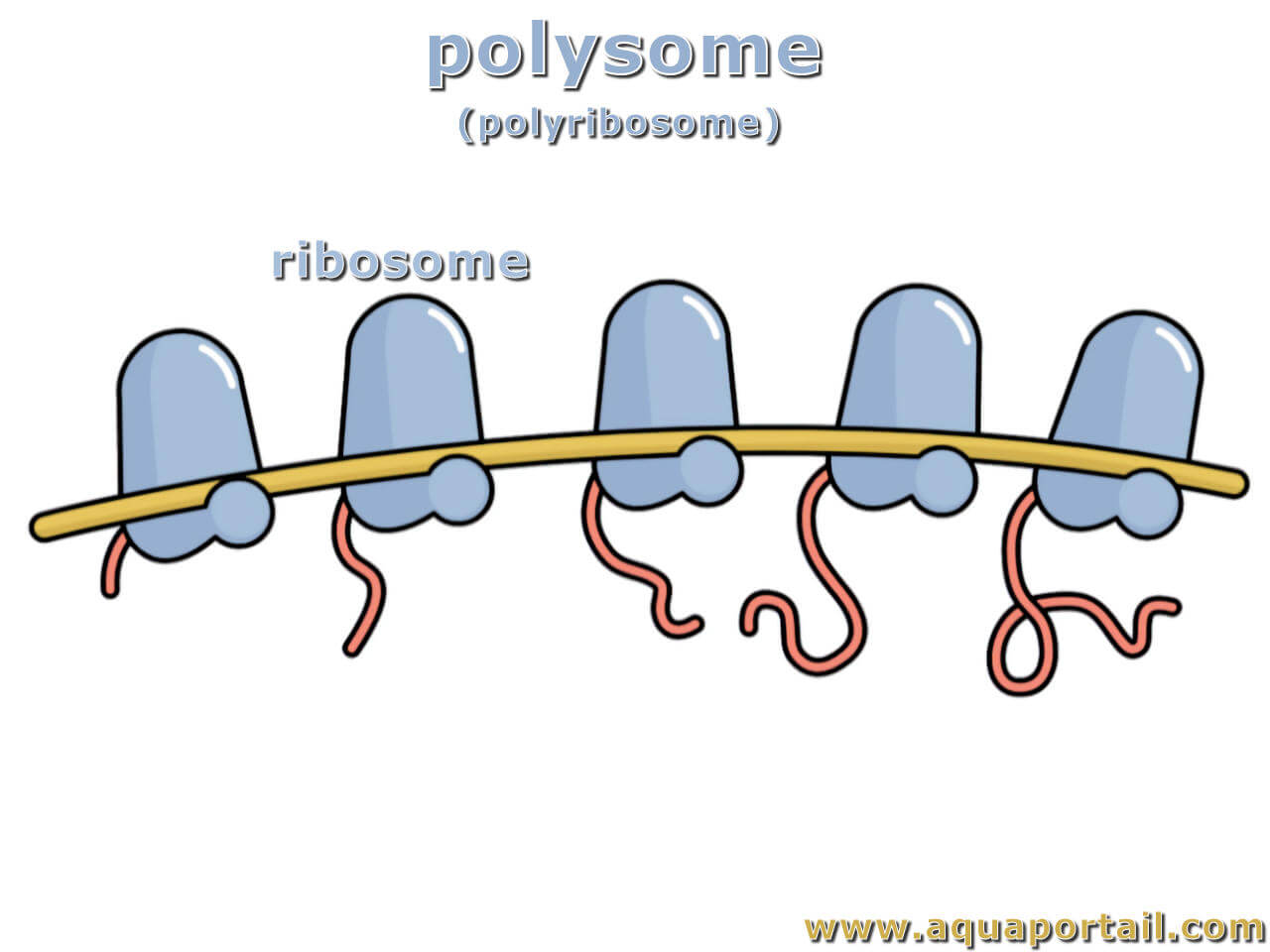
Polysomes
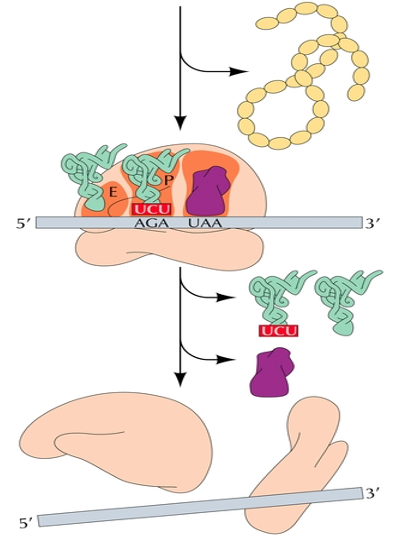
Polysomes
essentially "beads on a string", each "bead" is a ribosome and the "string" is the mRNA molecule.
Function:
Multiple ribosomes bind to the same mRNA, enabling the cell to produce many copies of the protein encoded by that mRNA quickly and efficiently.
How they form:
During translation, ribosomes sequentially load onto an mRNA molecule, forming a polysome.
Location:
found in both prokaryotic and eukaryotic cells, both in the cytoplasm and on the endoplasmic reticulum
Two classes of mutations:
Point mutations
Frameshift mutations
Point mutations & its types
Substitution mutations where one base is replaced with another base.
Three types of point mutation:
Silent mutation
Missense mutation
Nonsense mutation
A mutation that does not change the amino acid sequence of a protein
example: UUA change to UUG
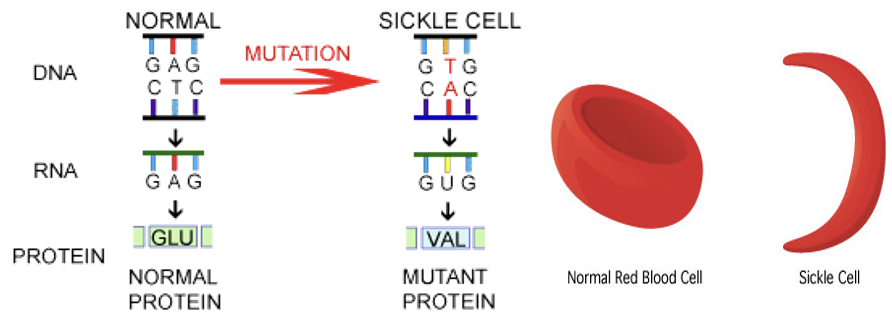
A mutation that switches one amino acid for another.
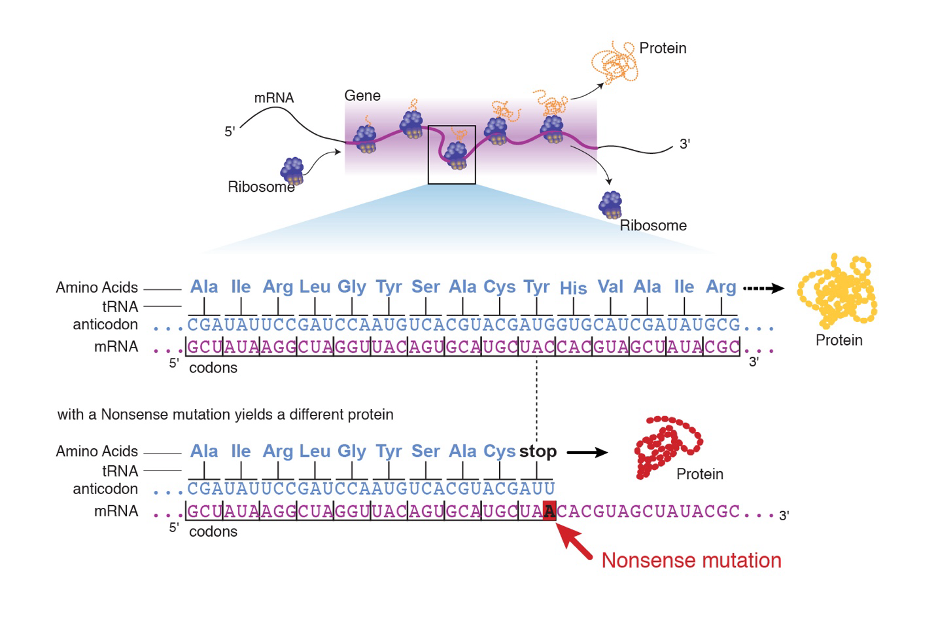
A mutation that results in an early stop codon in the protein.
Mutations that alter the entire reading frame of a mRNA sequence.
(nucleotides are read three nucleotides at a time)
Two types of frameshifts:
Insertion mutation
AUG-UCA-CUU-G
Deletion mutation
AUG-ACU-UG
Normal sequence:
AUG-CAC-UUG
Proteins that assist in the folding of other proteins.
They work by assisting other proteins to self-assemble, possibly by stabilizing the unfolded intermediates
example: proper folding might require both the carboxy and amino terminus. Chaperones maintain the protein until it is completely translated and ready to properly fold
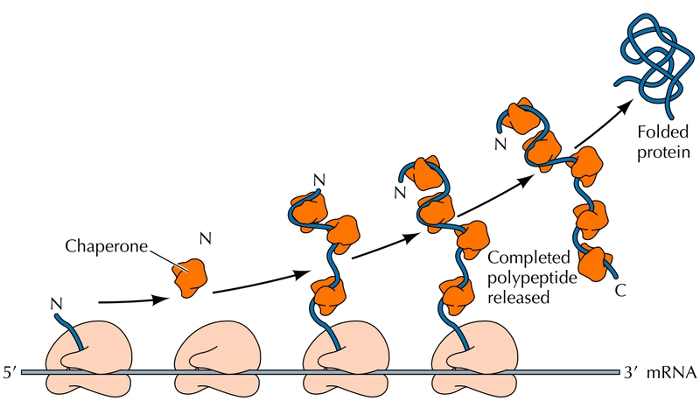
Protein cleavage
Cleavage of the polypeptide chain is important for maturation.
contains signal sequence and signal peptidase
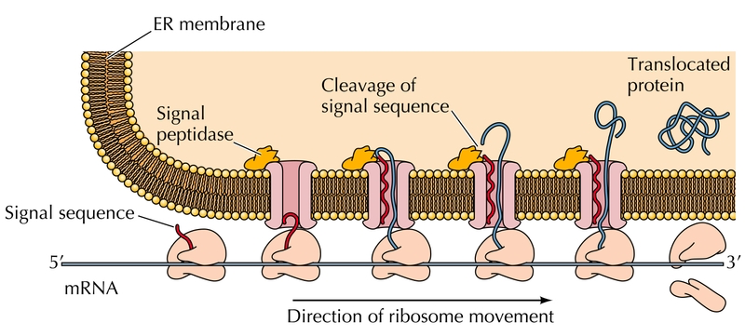
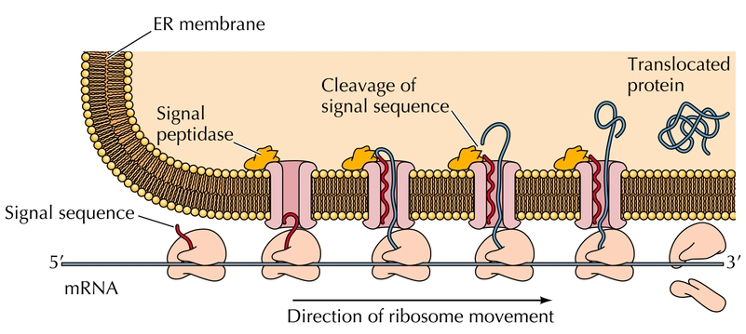
Signal sequence
20 amino acids at amino terminal that targets secreted proteins to the endoplasmic reticulum.
This sequence is hydrophobic and assists the protein through the membrane.
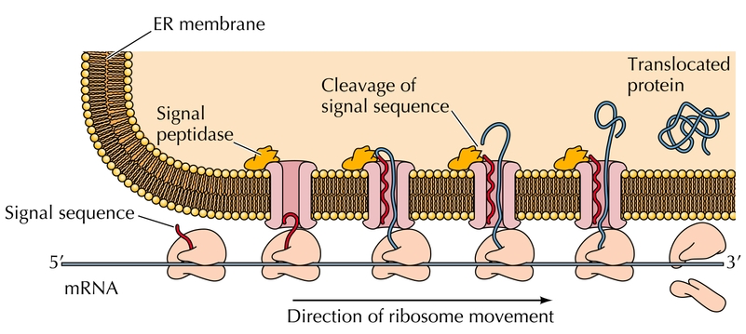
Signal peptidase
A sequence that allows a protein to enter the endoplasmic reticulum (ER).
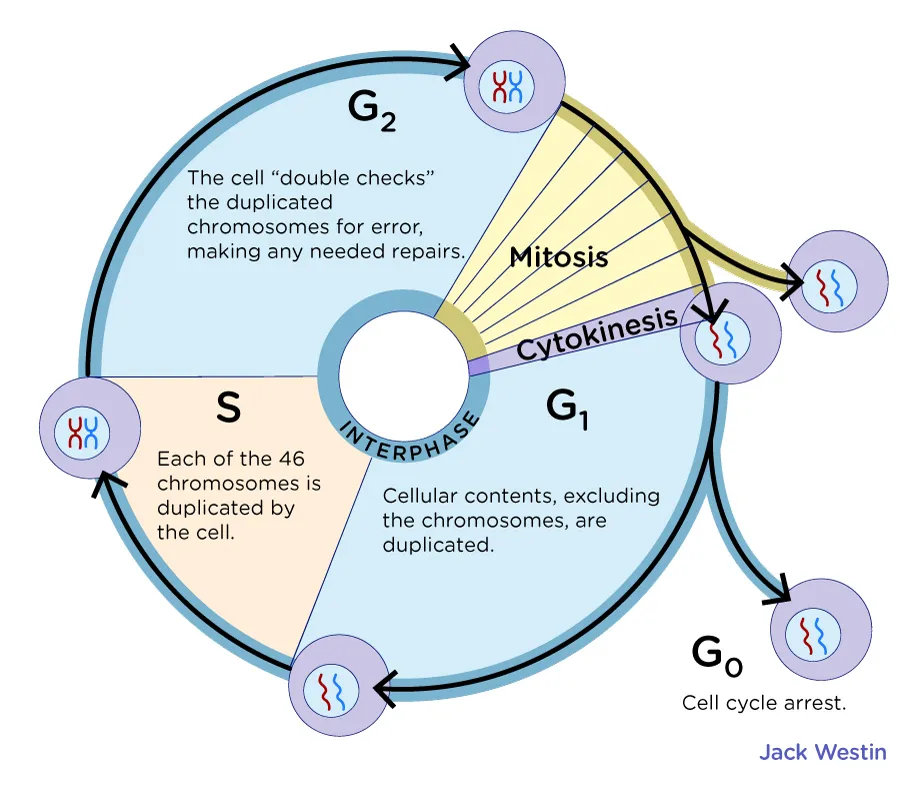
The cell cycle
Organism
^
Cells ← 2 cells
^
Organelle
^
Macromolecules
^
Atom
How many humans would it take to stretch our DNA from the moon to the Earth?
In a single human, we have enough DNA to stretch from the surface of the earth to the moon

Chromosomes
Offspring inherit chromosomes containing genetic material from parental generation.
Chromosomes are made of two sister chromatids.
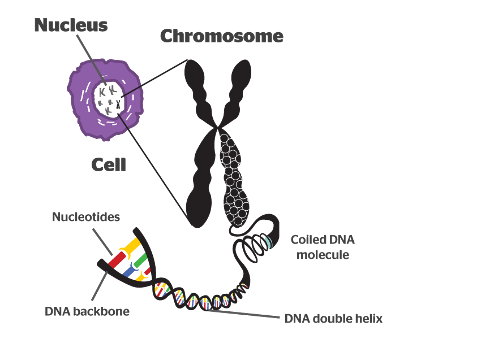
Sister chromatids
Made of chromatin
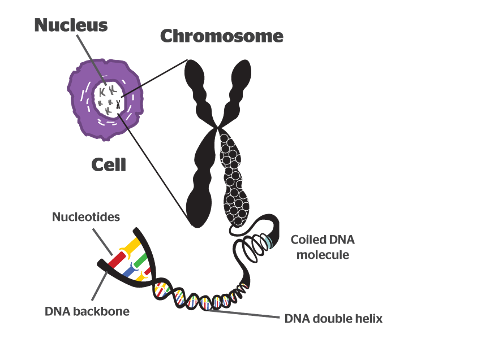
Chromatin
Made of histones and DNA
G1=
S =
each half of the X =
G2 =
G1= linear chromosome
S = after it replicates,
each half of the X = a sister chromatid but the whole X is a chromatid
G2 = X
X shape means that the chromosome has already divided
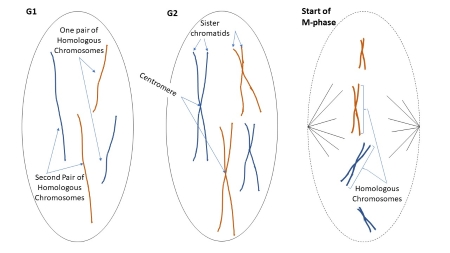
Chromosome pairs
In humans, there are 22 pairs of autosomes and 1 pair of sex chromosomes.
Y = male
Don’t have a Y = female
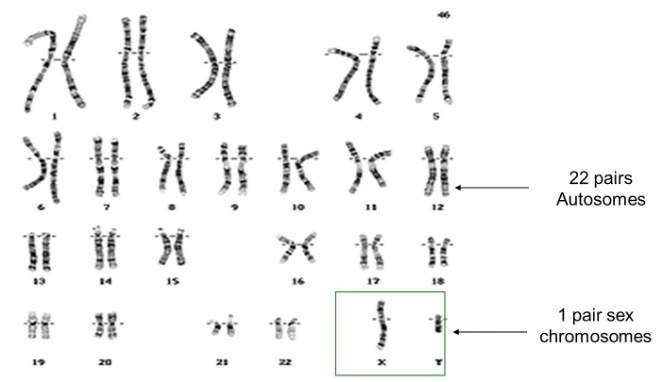
Chromosomes in Species
Chromosome number vary between species, the number of chromosomes within species remain consistent.
Genes are carried on chromosomes, what are genes?
Coding regions in DNA

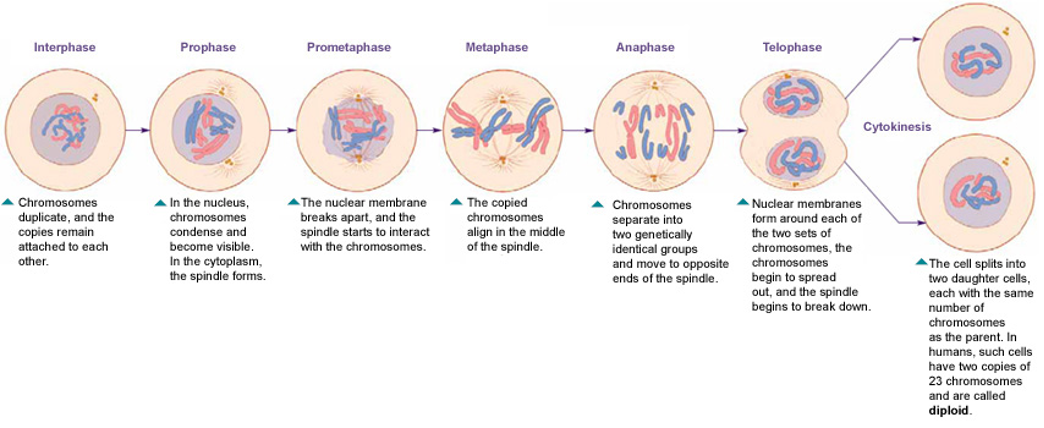
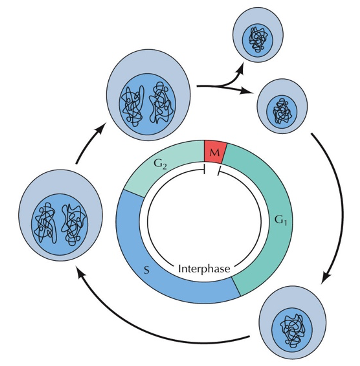
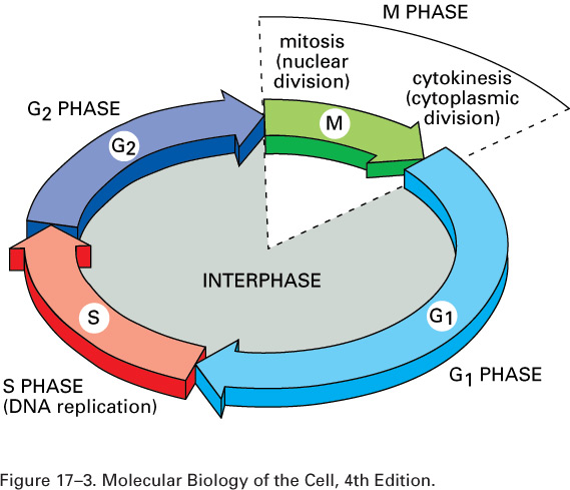
The phases of the cell cycle
Interphase
Mitosis (Diploid/somatic cells)
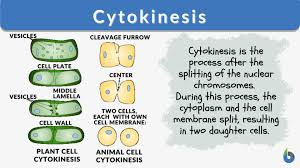
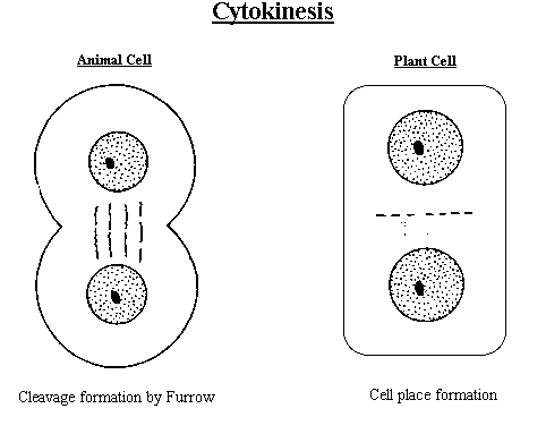
Cytokinesis
Length and division of cell cycle
The length of the cell cycle varies
Most adult cells divide slowly (once every few days or weeks)
Cancer cells and some normal cells (bone marrow, stomach) divide constantly.
Embryo cells also divide fast during cleavage
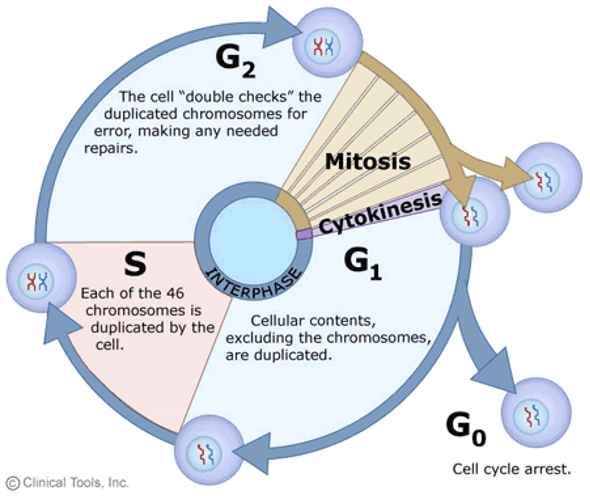
Interphase
G1 phase - Primary cell growth (cell organelles duplicated).
S - DNA synthesis
G2 - Secondary growth (centrosomes form)
G0 - Cell leaves the cell cycle and enters cell cycle arrest.
Organelles that organize microtubules during cell division.
Contains 2 centrioles which are small cylinder-shaped organelles that help form mitotic spindle.
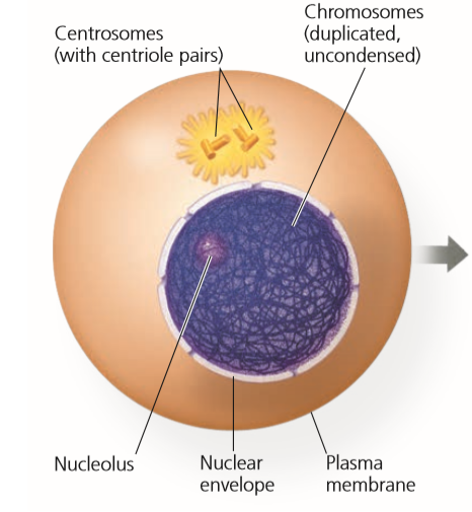
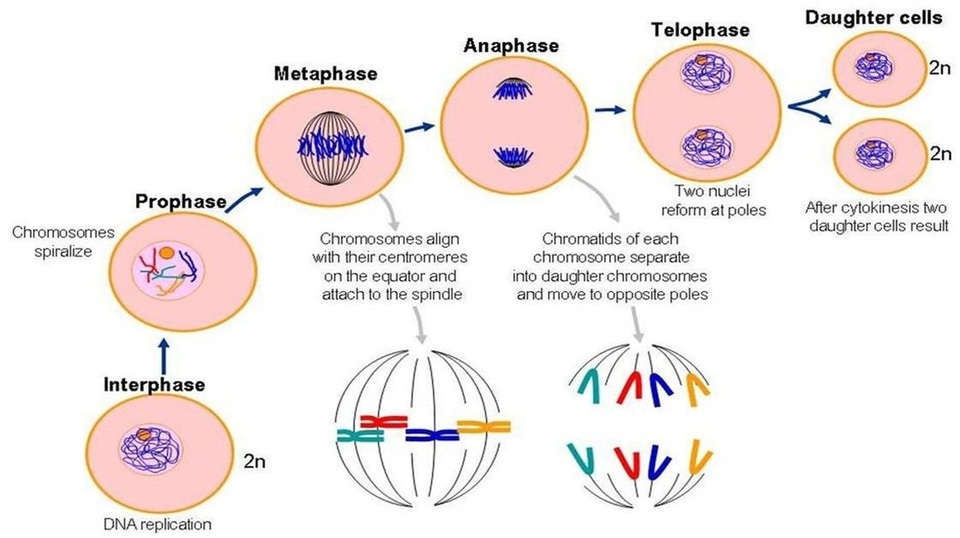
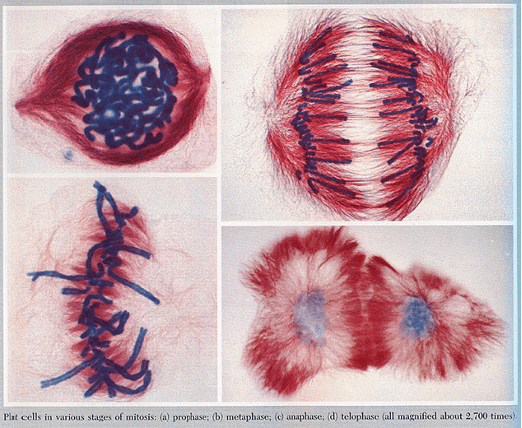
Process of cell division that includes stages:
prophase
prometaphase
metaphase
anaphase
telophase
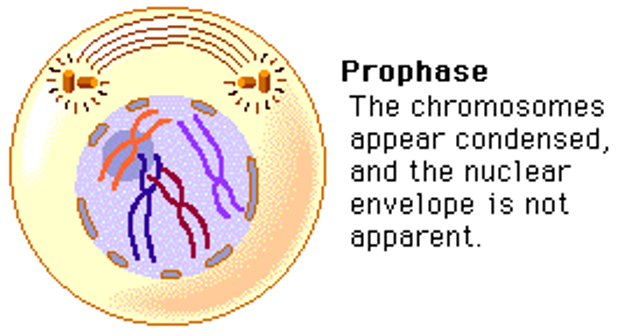
Prophase
Mitotic spindle forms, chromosomes condense, and nuclear envelope disappears.
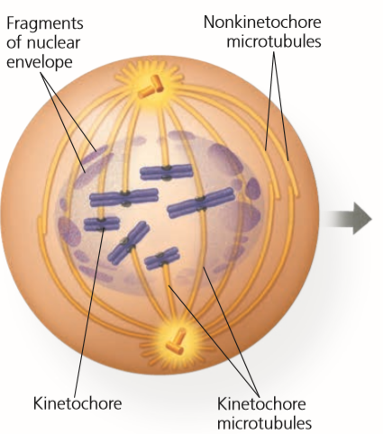
Prometaphase
Kinetochore forms at the centromere.
Centromere = Middle of the chromosome that connects the two arms of the chromosome
Some kinetochores attach to centromere while others interact with opposite poles of cell
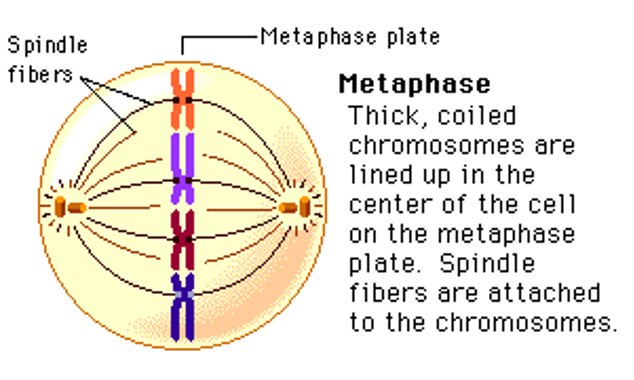
Metaphase
Centrosomes go to opposite poles of the cell.
Chromosomes line up along metaphase plate.
Sister chromatids are identical / no difference
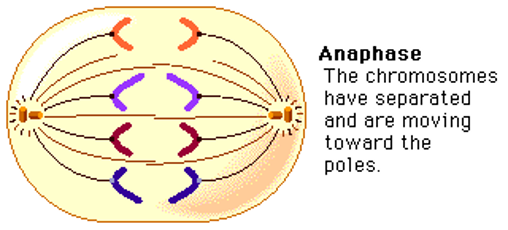
Anaphase
Cohesion protein that holds sister chromatids together are degraded and sister chromatids are separated to opposite poles.
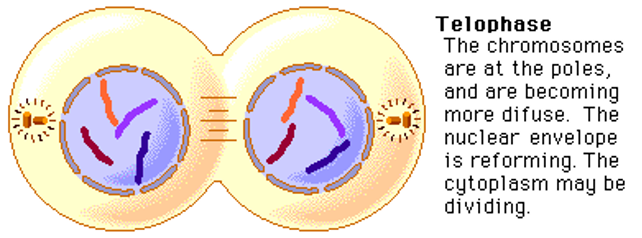
Telophase
Nuclear envelope reappears, chromosomes become less condense and microtubules disassemble.
Start unwrapping
Centrosome breaks apart
Cytokinesis for animal vs. plant cells
Animal cell forms cleavage furrow (actin filaments).
Plant cell forms cell plate from cell wall (vesicles).
Mitosis diagram order