Gen Lec 20: DNA Mutation + Repair
1/45
Earn XP
Description and Tags
TEST CONCEPTS: Recognize Mutation given a sequence and explain what the effect would be. Mutation Nomenclature (base 1/ base 2 -> base 1'/base 2'))
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
46 Terms
Mutation
Heritable change in genetic information
change from WT
Source of genetic var. that is necessary in population for evolution to happen
Wild Type
What is observed most often
Mutations Characterized by Size
Chromosomal, Point
Chromosomal mutations (def, e.g.)
large scale genetic alterations that affect structure / #
e.g. (deletions, inversions duplications, trans)
point mutations
small scale, single/few nucleotides are changed, added, or deleted
Missense, Nonsense, frameshift, silent
Mutations characterized by change to Nucleotide
Transitions, Transversions
Transition
Purine → Purine / Pyrimidine → Pyrimidine
More common/less significant change to chem structure
Transversions
Purine ↔ Pyrimidine
Significant change to chemical structure
Mutations characterized by Affects Mutation has on Protein
Missense, Nonsense, Silent, Frameshift
Missense
point mutation leads to amino acid change in protein
Nonsense
point mutation changes sense codon → nonsense codon
results in premature stop codon → truncated, usually nonfunctional protein
Sense vs. Nonsense Codon
Sense codes for amino acid; Nonsense Codes for stop
Silent Mutation
Point mutation changes codon, but amino acid stays the same
happens bc of degeneracy of code
Frameshift
addition/deletion of multiples 1 or 2 but not 3 base pairs
Shifts reading frame so that amino acid seq downstream of mutation is incorrect
Often puts nonsense codon in frame so final protein is truncated
Mutations characterized by where Mutations happen
Somatic Mutations, Germline Mutations
Somatic Mutations
Mutations in Body cells
Not heritable by progeny
Sometimes result in increased cell division → cancer
Not all cells in affected individual will have mutation, but daughter cells of a cell with a somatic mutation will have the mutation
Germline Mutations
in gametes
Heritable
All cells in affected individual have mutation
Mutations characterized by how they occur
Spontaneous mutations, Induced mutations
Spontaneous mutations (Definition + causes)
Occur during normal lifetime of a cell
Causes :
Errors made during DNA replication
Normal biochemistry in cell creates reactive Depurination, Deamination
Errors made during DNA replication
Incorrect base insertions likely due to wobble pairing
Strand slippage
Incorrect base insertions likely due to wobble pairing
If a replication error due to wobble pairing (e.g., insertion of T instead of C) is not corrected, a mismatch or bulge can form that escapes DNA proofreading
after a subsequent round of DNA replication → mutation becomes permanently established in genome
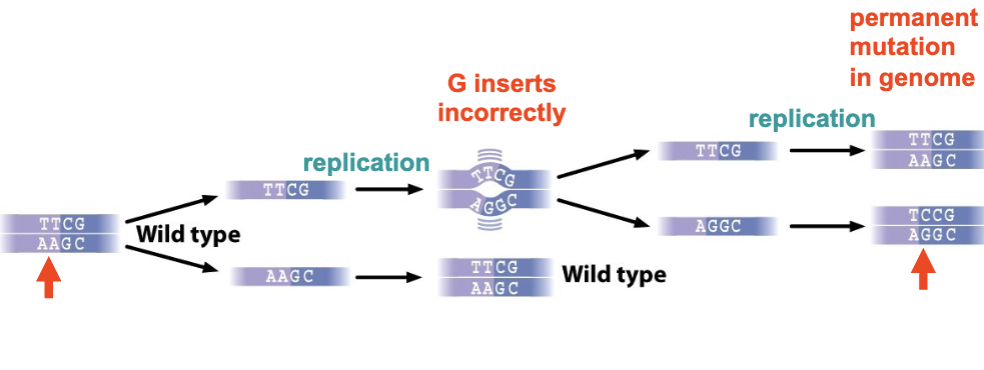
wobble pairing
H-bonding between non-Watson/Crick bases
Frameshift mutations due to strand slippage
Strand slippage occurs during DNA replication at repetitive sequences when DNA polymerase “loses count“ and misaligns the strands
extra bases are added → newly synthesized strand loops out → insertion
bases are skipped → template strand loops out → deletion
Depurination
Loss of purine base AG
sugar phosphate backbone stays intact
Normally repaired (Base excision), but when DNA Pol gets to site w/o base, it doesn’t know what to do and often inserts A
→ subsequent round of DNA rep → permanent change in genome

Deamination (Def and 2 cases)
Loss of amino group from a base
(Can You Anak)
Amino group lost from cytosine → yields uracil → pairs w/ A during replication (C-N=U+A)
G/C → A/T after 2 rounds of DNA replication (transition)
Amino group lost from Adenine → hypoxanthine → pairs w/ C during replication (A-N = HA+C)
A/T —> G/C after 2 rounds of DNA replication (transition)
Induced mutations / (mutagen types)
results from exposure to external agent (mutagen)
base analogues, base modifiers, intercalating agents, UV light
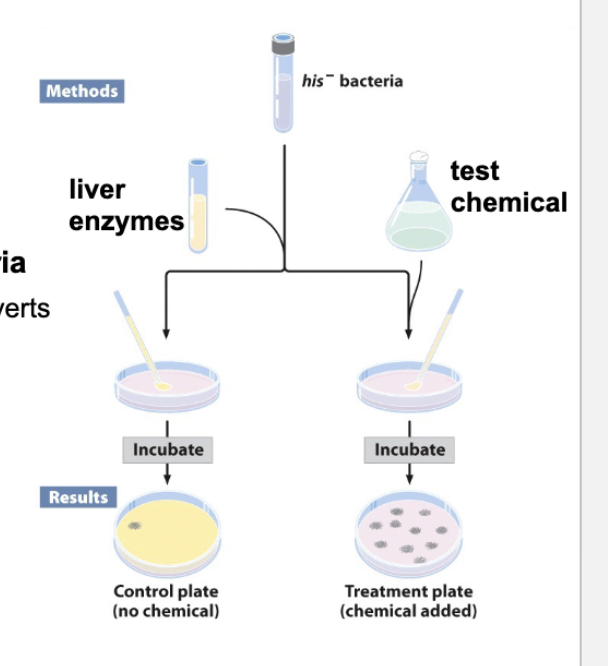
AMES Test (Purpose, Method)
Used to identify mutagens
uses mutant bacterial strain (No DNA repair systems + cannot make Histidine)
Expose bacteria to chemical w/ and w/o Liver enzyme (to test if metabolized chemical is a mutagen)
Plate bacteria treated w/ chemical, chemical + liver enzyme, and untreated control on minimal media
Growth → His+ mutation (bacteria has been mutated and reverted back to WT)
Base analogues
Chemical that looks like a base
can be incorporated into DNA during S phase
ionization will change basepairing properties during subsequent rounds of DNA replication
Base modifiers
add/remove chemical groups from a base so that it will not pair properly when it is template during DNA replication
Intercalating agents
flat planar molecules that intercalate between stacked bases in DNA
Contorts helical structure → insertions/deletions during DNA replication
UV Light
Cause adjacent pyrimidines to covalently bond together → they will NOT bp with complementary DNA strand during replication
Epistasis
Genotype at 1 locus mask gene expression at another locus
Intergenic Suppression
Individual w/ mutation is double mutant
Mutation 1 → causes muation
Mutation 2 (at another site)→ suppresses effect → WT
Intergenic suppression via suppressor tRNA
Nonsense Mutation 1 Eye Color Gene → Truncated Protein
Suppressor Mutation 2 in tRNA Gene → Mut tRNA has anticodon that can base pair with stop codon → Full length of protein
Intergenic suppression via protein–protein interaction
Enzyme that needs multiple subunits (A/B) to function
Missense Mut 1 (Subunit A gene) → Subunit A changes and can no longer bind Subunit B
Suppressor Mut 2 (Subunit B gene) → Subunit B changes and can bind with A
Intragenic Suppresion (def + 2 ecamples)
Second mutation in a SAME gene restores (partially or fully) the phenotype caused by the first mutation
E.g. mutations in same codon, mutations in different codons
Intragenic Suppresion: Both mutations are in a single codon
1st point mut: Leu → Phe
2nd point mut: Phe → Leu
occurs because of redundancy in genetic code
Intragenic Suppresion: Mutations are in different codons of the same gene (± example)
aa1 and aa2 must interact tfor protein to fold correctly
1st point mut: aa1 (-) charge → (+) charge
2nd point mut: aa2 (+) charge → (-) charge
changes restore original proteins structure
DNA Repair Mechanisms
Require 2 strands of DNA (mutated; nonmutated strand = template)
Multiple ways to do the same thing
Direct Repair (Def + 2 examples)
Reverse whatever just happened
Mutation: Thymine dimers caused by UV light
Photolyase, activated by light destroys covalent bonds between Ts so they can create H-bonds
Base is Methylated
Transferase enzymes remove alkyl + methyl gr
2 types of excision repair
Nucleotide excision repair, Base excision repair
Nucleotide excision repair
Cells are not actively dividing
repair proteins recognize distortion in helix as an error
Endonuclease cleave the phosphodiester bond on either side of distortion → remove that piece of DNA
Gap is filled by DNA Pol
Nick is sealed by DNA
Base Excision Repair
involves initial removal of base
DNA Glycolase removes modified base
DNA endonuclease nicks DNA and removes sugar
DNA polymerase adds new nucleotide
Nick is sealed by DNA ligase
Mismatch repair
Corrects mismatches that happen during replication
new strands (w/ mistake) are distinguished from old strains (no mistake) bc more A’s are methylated on old strand than new strand
enzymes look for distortions in DNA
section w/ mutation is excised
DNA Pol I fills in gap
DNA ligase seals nick
Repair Systems that repair DNA mutations in cells that are not actively dividing
Mismatch, Direct, Base, Nucleotide excision
Repair Systems that repair DNA mutations in cells that are actively dividing
Homologous recombination / Nonhomologous end joining