MB&B Study Guide ~ Exam 3 ~ Post Transcriptional / Post Translational Control of Gene e
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
22 Terms
Does the rate of transcription of a gene correlate perfectly with the amount of protein produced from that gene? Why or why not?
The rate at which a gene is transcribed into mRNA doesn’t always match the amount of protein that ends up being produced from that mRNA.
This is because different mRNAs have a wide range of stability, meaning some mRNAs break down quickly while others are more stable and last longer. So, even if there is a high rate of transcription and a lot of mRNA is made, if it breaks down quickly there may not be enough time to produce a lot of protein from the mRNA transcripts.
mRNA Half Lives
The half-life of mRNA is how long it takes for half of a population of mRNA molecules to break down
Not all mRNAs decay at the same rate
How does half life differ in response to temporary changes in the environment? Why?
mRNAs that are produced in response to temporary changes in the environment usually have shorter half-lives.
If a cell needs to react quickly to heat or stress, it will make mRNAs that break down fast so that it can stop making those proteins when they’re no longer needed
Can have different stabilities in different cell types
How is mRNA stability determined?
mRNA stability can be built into their sequences or change based on the conditions of the cell
“Hard wired or regulated”
mRNA with AU rich elements
tend to have shorter half lives. These sequences are found in the 3’ UTR of many mRNAs. Experimental proof:
B-globin gene normally has ten hours of stability
When inserting the AT rich sequence of the GM-CSF gene, the produced mRNA has only 1-2 hours of stability.
Inserting a GC rich sequence doesn’t change the half-life of b-globin mRNA
This is because certain proteins can bind to AU rich sequences and affect how quickly mRNA breaks down.
Discuss mechanisms besides transcription that can affect the production of proteins.
?
How does iron enter the cell?
Iron entry: Iron in the bloodstream is transported by a protein called transferrin. Cells have transferrin receptors on their surface that recognize transferrin proteins. If a cell needs iron, transferrin binds to iron, allowing iron to enter the cell through the transferrin receptors.
How is iron stored in the cell?
Iron storage: Once inside the cell, iron can be stored by a protein called ferritin. Ferritin helps protect the cell from excess iron which can be harmful due to its oxidative properties.
If there is too much iron, ferritin will collect it and more ferritin will be produced
If there is too little iron, the cell will increase the uptake of iron through transferrin receptors
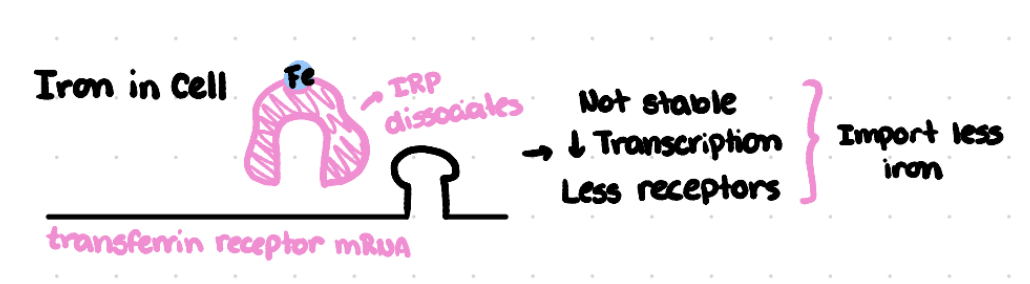
How does the presence of iron regulate the half-life of the transferrin receptor mRNA?
The presence or absence of iron regulates the half-life of transferrin receptor mRNA using the iron response protein.
When iron is available, it binds to IRP, causing IRP to dissociate from the transferrin receptor mRNA.
This leads to a reduction in the half-life of mRNA meaning it is degraded more quickly.
Less transferrin receptor protein is produced reducing the cell’s ability to take in iron.
How does the absence of iron regulate the half-life of the transferrin receptor mRNA?
When iron levels are low, IRP binds to a specific sequence in the UTR of transferrin receptor mRNA
This binding stabilizes the mRNA which increases its half life and promotes translation into transferrin receptor proteins.
The cell can import more iron because there are more transferrin receptor proteins

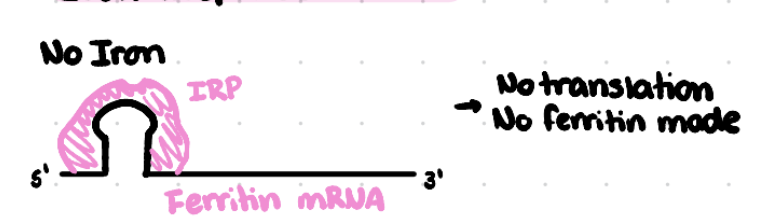
How does the Iron Response Protein (IRP) regulate translation of the ferritin mRNA in absence of iron?
IRP binds to iron response element (IRE) located in the 5’ UTR of ferritin mRNA
This inhibits translation of ferritin protein — production of ferritin is reduced, less ferritin in the cell.
This allows iron to be available for cellular needs instead of being taken up by ferritin
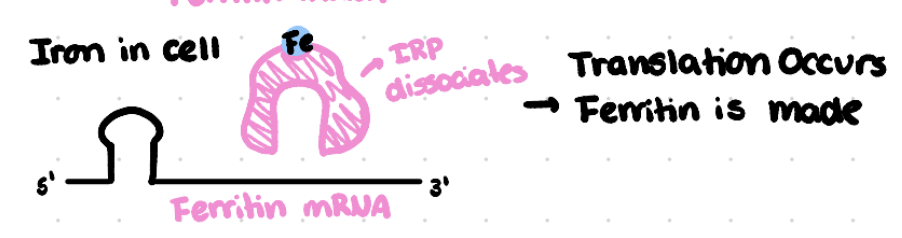
How does the Iron Response Protein (IRP) regulate translation of the ferritin mRNA in presence of iron?
Iron binds to IRP which causes it to dissociate from the IRE on the ferritin mRNA
This allows ferritin mRNA to be translated leading to more ferritin protein.
Ferritin is then able to take up the excess iron, protecting the cell.
Describe RNA interference (RNAi)
RNA Interference, or RNAi, is a defense mechanism cells use against viruses / viral infections by degrading viral RNA and regulating gene expression.
Induced by long stretches of dsRNA
Our cells typically do not have long stretches of dsRNA, so it signals we are being invaded by a virus
How is double stranded RNA processed to form siRNAs?
dsRNA is recognized by cell machinery and cleaved into small pieces. It then binds to and degrades mRNA molecules with matching sequences, which silences the gene expression associated with that mRNA. This is good if the mRNA is a viral mRNA.
?
Understand the roles of dicer, RISC, and argonaute.
Dicer ~ Recognizes / cleaves dsRNA into smaller pieces known as siRNAs which are 20-25 nucleotides long
RISC ~ Binds to siRNAs and discards one strand. Then uses the remaining strand to look for complementary RNA sequences (like viral RNA).
If the half of siRNA base pairs with a viral RNA it is able to get them degraded.
Argonaute ~ When siRNA base pairs perfectly/near perfectly to an RNA, argonaute cleaves the RNA (so it is no longer functional). When siRNA base pairs pretty well/partially, the argonaute will sit on the strand and stop it from being translated.
In both cases the mRNA transcript will be degraded
Describe the regulation of gene expression by microRNAs (miRNAs).
miRNA / microRNAs ~ small RNA molecules that help regulate gene expression by binding to mRNAs which leads to either:
Degradation of mRNA ~ mRNA is broken down and cannot be used to make protein
Inhibition of translation ~ miRNA prevents mRNA from being turned into a protein silencing the gene
miRNAs are naturally produced by the cell
pre-miRNA are transcribed, and RNA-RNA base pairing gives them a hairpin structure
Endonuclease Drosha cleaves them
Undergo a process similar to siRNA
pre-miRNA is loaded onto dicer
Dicer cleaves
miRNA binds to RISC and looks for mRNA
Near perfect complementarity → argonaute cleaves the mRNA. Partial complementarity → argonaute will sit on the strand and stop it from being translated.
In both cases the mRNA transcript will be degraded because it’s not being translated
Understand the role of RISC (the RNA-induced silencing complex) in using microRNAs to recognize target mRNAs and either degrade them or inhibit their translation.
When miRNAs are formed, they get incorporated into RISC
RISC uses miRNA to find matching mRNA in cell
miRNA in RISC binds to complementary target mRNA
Perfect/near perfect base pairing → RISC cleaves mRNA leading to degradation
Partial base pairing → RISC prevents the mRNA from being translated into protein by sitting on it
How can the miRNA/RNAi pathways be used as tools to repress expression of specific genes in cells you are studying?
Researchers can design miRNAs or siRNAs that are complementary to specific mRNAs of interest. By introducing these cells, scientists can silence the expression of specific genes.
When synthetic siRNA or miRNA is introduced into the cell:
It will get incorporated into the RISC complex just like natural miRNAs or siRNAs.
RISC uses synthetic RNA to find and bind to the target mRNA
RISC either degrades the mRNA or inhibits its translation
This is helpful for studying gene function, because by silencing a specific gene researchers can observe the effects the gene has on the cell / organism helping to understand the role of that gene.
What is “codon bias”?
Different codons that code for the same amino acid are not used with equal frequency
Even though the genetic code is redundant (multiple codons can code for the same amino acid) certain codons are still preferred over others, which changes depending on organism / tissue
Species evolutionarily choose to use one codon more than the other when both coding for the same amino acid - habitual.
As a result they have more of those tRNAs
Can impact protein production – two mRNAs that encode the same protein (using different codons) may result in different levels of protein expression due to differences in translation efficiency
How can the abundance of specific tRNAs affect the efficiency of translation?
The abundance of specific tRNAs affects translation efficiency because tRNAs match codons on mRNA to the correct amino acids. If a tRNA for a codon is abundant, the translation will be fast and efficient. If the tRNA is rare, translation will slow down as the ribosome has to wait for the right tRNA to arrive.
Since different codons can code for the same amino acid, those recognized by abundant tRNAs are translated more efficiently than codons recognized by rare tRNAs. This is why mRNAs using common codons will produce more protein while those with rare codons translate less efficiently.
How is protein stability related to its N-terminal amino acid? (Don’t have to know which N-terminal residues are stabilizing or destabilizing, just know the concept).
Protein stability is influenced by its N-terminal amino acid.
N-terminal amino acid is revealed after the initial methionine is cleaved during translation (in 2/3rds of proteins).
The identity of the N-terminal amino acid will affect how long the protein will last in the cell. Some N-terminal amino acids are recognized by ubiquitin ligases which tag the protein for degradation.
Proteins with destabilizing N-terminal amino acids are targeted for quicker degradation
Proteins with stabilizing N-terminal amino acids lead to a longer protein lifespan
Describe the role of ubiquitination in controlling the stability of proteins.
Ubiquitin ~ Controls protein stability by tagging proteins for degradation
Ubiquitin is a small protein that covalently attaches to lysine residues on target proteins. Additional ubiquitin molecules then attach to the lysine residues on each other to form a polyubiquitin chain
Polyubiquitin chains signal the proteasome, a protein-degrading complex, to recognize, unfold, and break down the tagged protein.
Ubiquitination can target misfolded or damaged proteins nonspecifically, or it can be more specific with E3 ubiquitin ligases recognizing particular proteins for degradation. This process allows cells to regulate protein levels and remove defective proteins