Genetics Week 5 Pt. 2
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
11 Terms
What if we consider three genes that are all located on the same chromosome? If m, w, and y are all located on the X chromosome, what can we say about the relative distance between them?
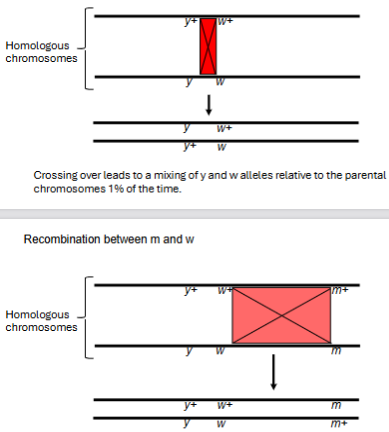
m and w 32.8% recombinants
w and y 1% recombinants
Recombination between y and w
Crossing over leads to a mixing of y and w alleles relative to the parental chromosomes 1% of the time.
Crossing over leads to a mixing of m and w alleles relative to the parental chromosomes 32.8% of the time.
Mapping of genes + specific points + how to construct gene map
Gene/chromosome maps assign a gene to a specific location on a chromosome. This specific location is called a locus.
Knowing where a chromosome a gene is is important for isolating the DNA and understanding its function and regulation.
Using multiple crossing experiments tracking a few genes at a time, a gene map can be constructed from observed recombination frequencies.
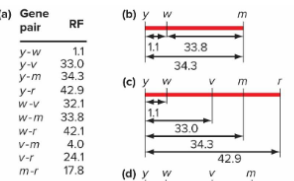
Mapping genes by comparisons of two-point crosses (example with m, w, and y)
Comparing different two-point crosses helps establish gene positions.
m and w 32.8% recombinants
w and y 1% recombinants
m and y ?
Adding the distance between y and m allows us to complete the gene map.
Limitations of two-point crosses
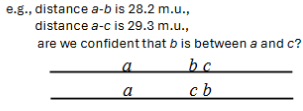
Difficult to determine the correct order of genes that are very close together.
Actual distances on the map do not always add up for genes that are far apart.
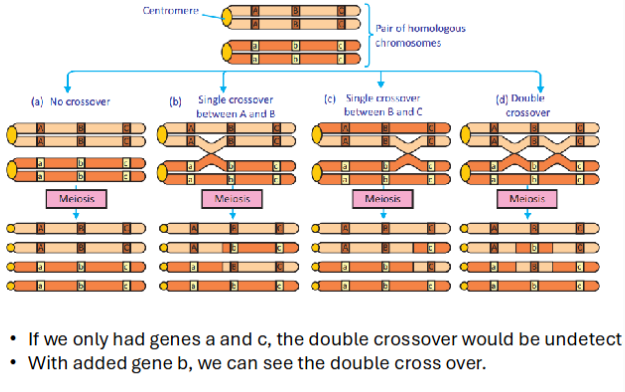
How do double crossover events mess up mapping distances?
Double crossover events are undetectable because they look like the parental chromosome configuration. This results in more individuals in the parental category and fewer in the recombinant category.
Fewer individuals in the recombinant category = genes estimated to be closer together than they actually are.
Large distance = likelihood of double crossover event between two genes relatively high = fewer recombinants detected than there should have been = underestimate of distance between y and r.
Three-point crosses + example with genes a, b, and c
Simultaneous analysis of three marker genes.
Faster, more accurate.
Allows correction for double crossovers.
Three-point cross example + calculations + CORRECTION
Cross heterozygote for three genes against homozygous recessive for the three genes. 8 pheno/geno classes
Two most frequent classes are parental.
Next two most frequent are a result of crossing over between the most distant genes.
Next two most frequent are a result of crossing over between the two closest genes.
Two least frequent are the result of a double crossover.
Step 1: determine gene order by calculating distances between each pair of genes.
Distance between vg and b:
- Parental types are vg b and vg+ b+.
- Find progeny that have combinations other than vg b and vg+ b+.
- Calculate recombination frequency
(252 + 241 + 131 + 118) / (4197) = 0.177 = 17.7% = 17.7 m.u.
Distance between vg and pr:
(252 + 241 + 13 + 9) / (4197) = 0.123 = 12.3% = 12.3 m.u.
Distance between b and pr:
(131 + 118 + 13 + 9) / (4197) = 0.065 = 6.5% = 6.4 m.u.
Based on the calculated distances, the gene order should be (vg - pr - b).
Correction:
13 and 9 are not added when ignoring pr as they are the parental types in terms of vg and b. Looking at the last two classes, they are also recombinant relative to vg and b, we just couldn’t see it because it appears parental.
Corrected calculation for distance between vg and b:
(252 + 241 + 131 + 118 + 13 + 13 + 9 + 9) / (4197) = 0.187 = 18.7% = 18.7 m.u.
Double crossovers are added twice because they involve two separate recombination events, both of which affect the distance between the outer genes, and would otherwise be missed in the original calculation.
Just by looking at the test cross data, which gene is in the middle? + Construct the gene map
Find parental genos:
q+ op+ + c+
q + op + c
Find double crossover genos:
q + op + c+
q+ + op+ + c
Only gene that is different is c. C is the gene in the middle. (2 unchanged + 1 changed)
Distance between q and c: 13.09 m.u.
Distance between c and op: 6.91 m.u.
Distance between q and op: 18.65 m.u.
CORRECTED distance between q and op: 20 m.u.


Examples of chromosomes located far apart on the same chromosome: (Are genes a and b linked?)
/ notation tells us they are on the same chromosome.
Frequency of each progeny class is close to 1:1:1:1.
If they are on the same chromosome, the data tells us they are very far apart.
These genes belong to the same linkage group, but they effectively assort independently.

X-linked data from males of a cross. What’s the genotype of the F1 female? What’s the gene order?
Parental genotypes are:
v b+ p+ and v+ b p. Thus genotype of mother is v b+ p+ / v+ b p
To find the middle genotype compare parental to double cross:
Parental:
v b+ p+
v+ b p
Double crossover:
v b+ p
v+ b p+
Only gene that is different is p. Gene in the middle is p.
Gene order is b p v.
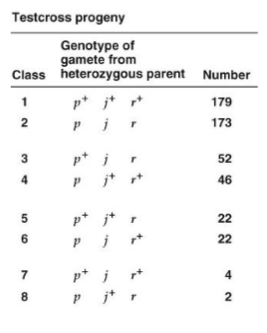
What is the parental class?
Which gene is in the middle?
What are the map distances?
Parental class is class 1 and 2.
Gene in the middle is j.
Distance between j and p: 20.8 m.u.
Distance between r and p: 30.8 m.u.
Distance between j and r: 10 m.u.