BIO 120.12 | Module 2: DNA Repair Mechanisms
1/106
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
107 Terms
Repairs UV damage (Thymine dimers)
Photoreactivation, nucleotide excision repair
First DNA repair mechanism discovered
Photoreactivation
First DNA repair mechanism discovered
UV damage (thymine dimers)
Photoreactivation
Light-repair enzyme
Activated by visible light (300 - 600 nm)
Folic acid (cofactor) absorbs light
Uses energy from absorbed light to perform cleavage of T-T (thymine) dimers
Photolyase
Enzyme used in photoreactivation to cleave thymine dimers
Photolyase
At what wavelengths is photolyase activated
Visible light (300-600 nm)
Cofactor of photolyase responsible for absorbing light
Folic acid
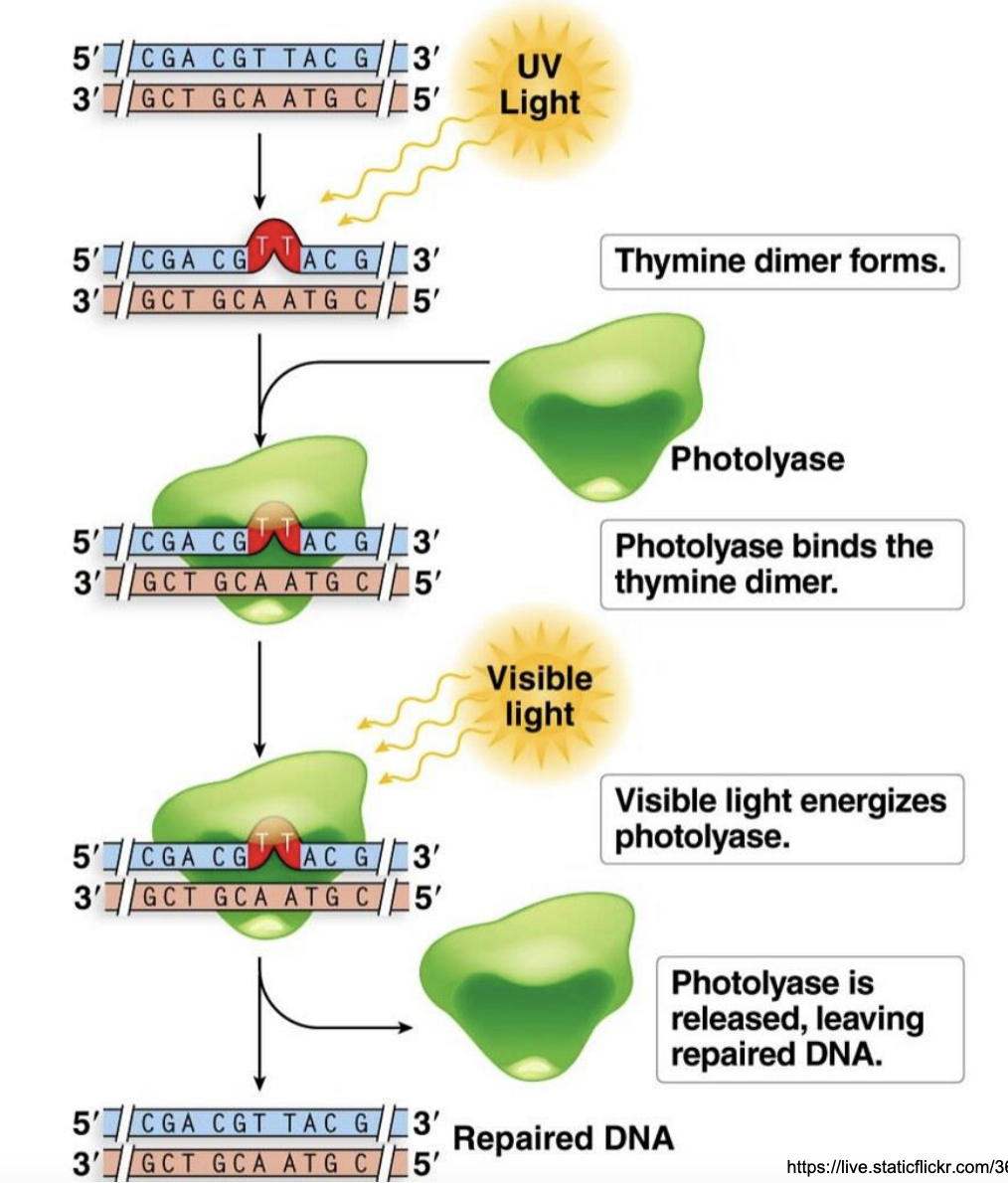
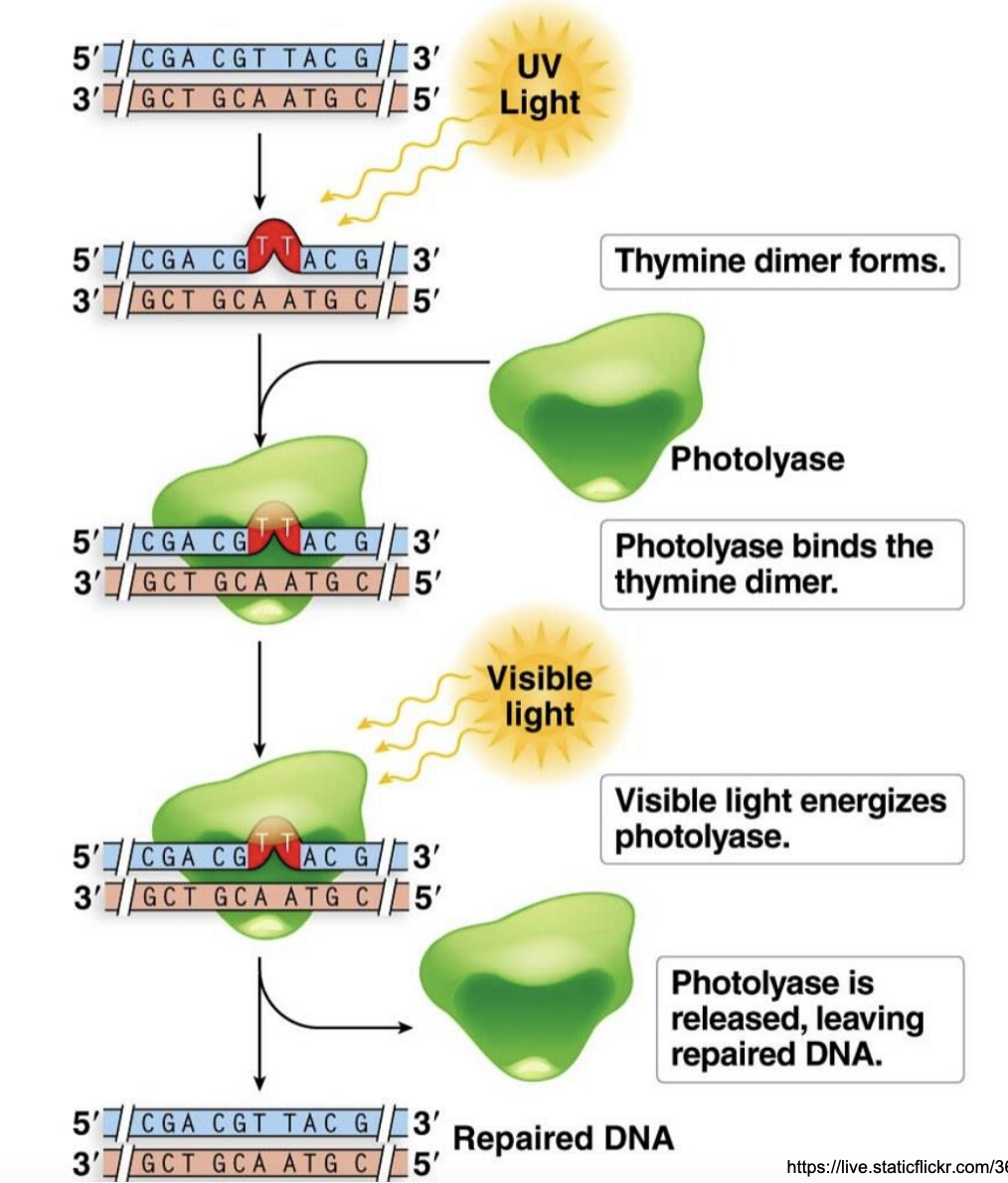
Explain photoreactivation
Exposure to UV causes formation of thymine dimer in dsDNA
Photolyase binds to thymine dimer
Visible light activates photolyase
Vis light activates folic acid cofactor
Energy from absorbed visible light is used by photolyase to repair thymine dimer
Photolyase released, leaving repaired DNA
Allows for localized excision and resynthesis of nucleotides at site of mismatch
Mismatch excision repair
Large structural distortion or chemical modification in the DNA, caused by the attachment of large molecules or chemical groups to the DNA bases
Bulky lesion
Repairs replicative errors within newly synthesized DNA
Mismatch excision repair
Repairs UV-induced damage, bulky lesions, mutations from other causes
Nucleotide excision repair
What type of bond is formed (and cleaved by photolyase) between T-T dimers
Glycosidic bond
T/F: Photolyase is inactive without visible light
TRUE
Methyl-directed post-replication repair system
Mismatch excision repair
T/F: Photolyase only repairs pyrimidine dimers
TRUE
Enzymes that cut DNA at internal sites rather than at ends
Endonuclease
Enzymes that remove nucleotides from the end of DNA, cutting 1 nucleotide at a time
Exonuclease
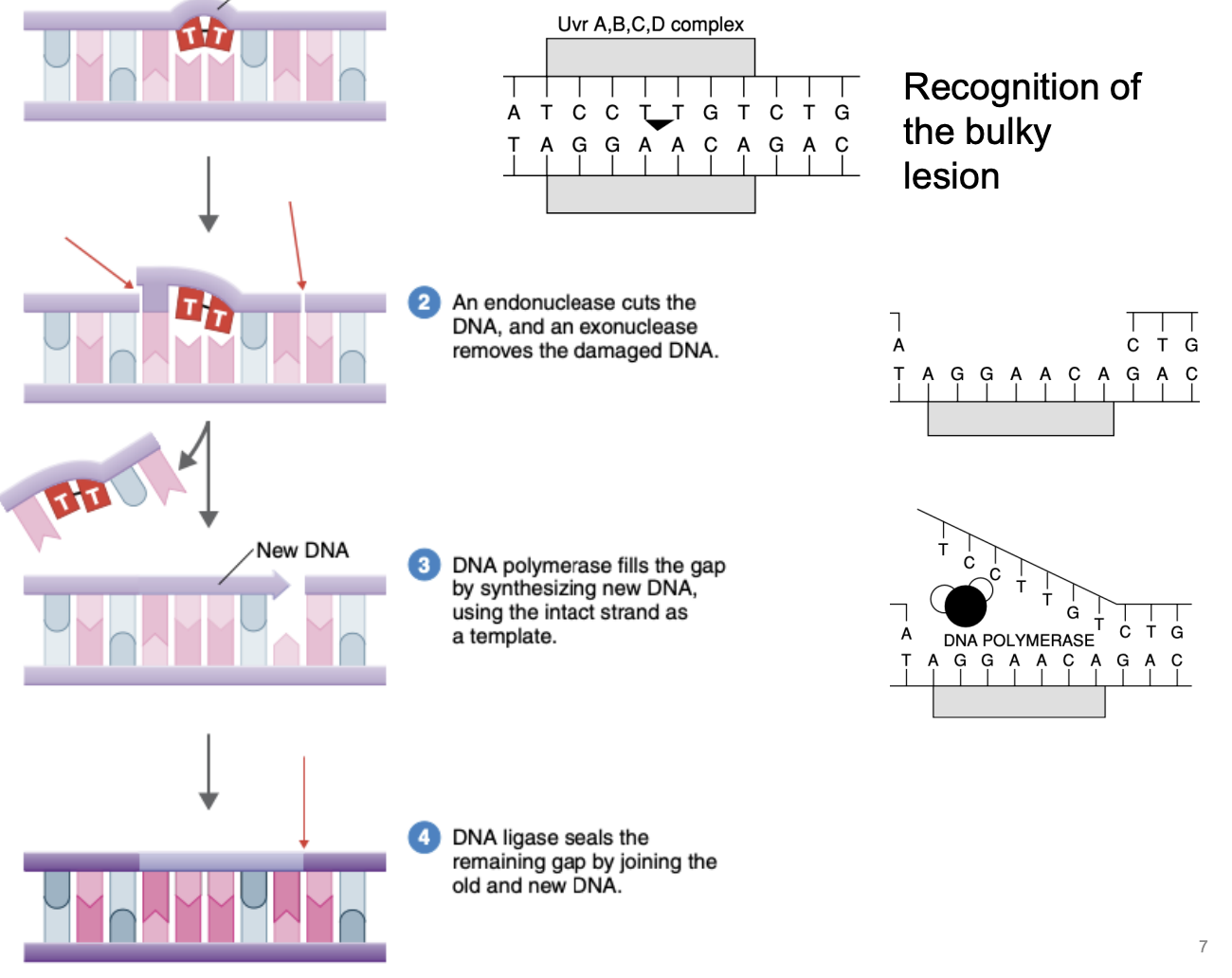
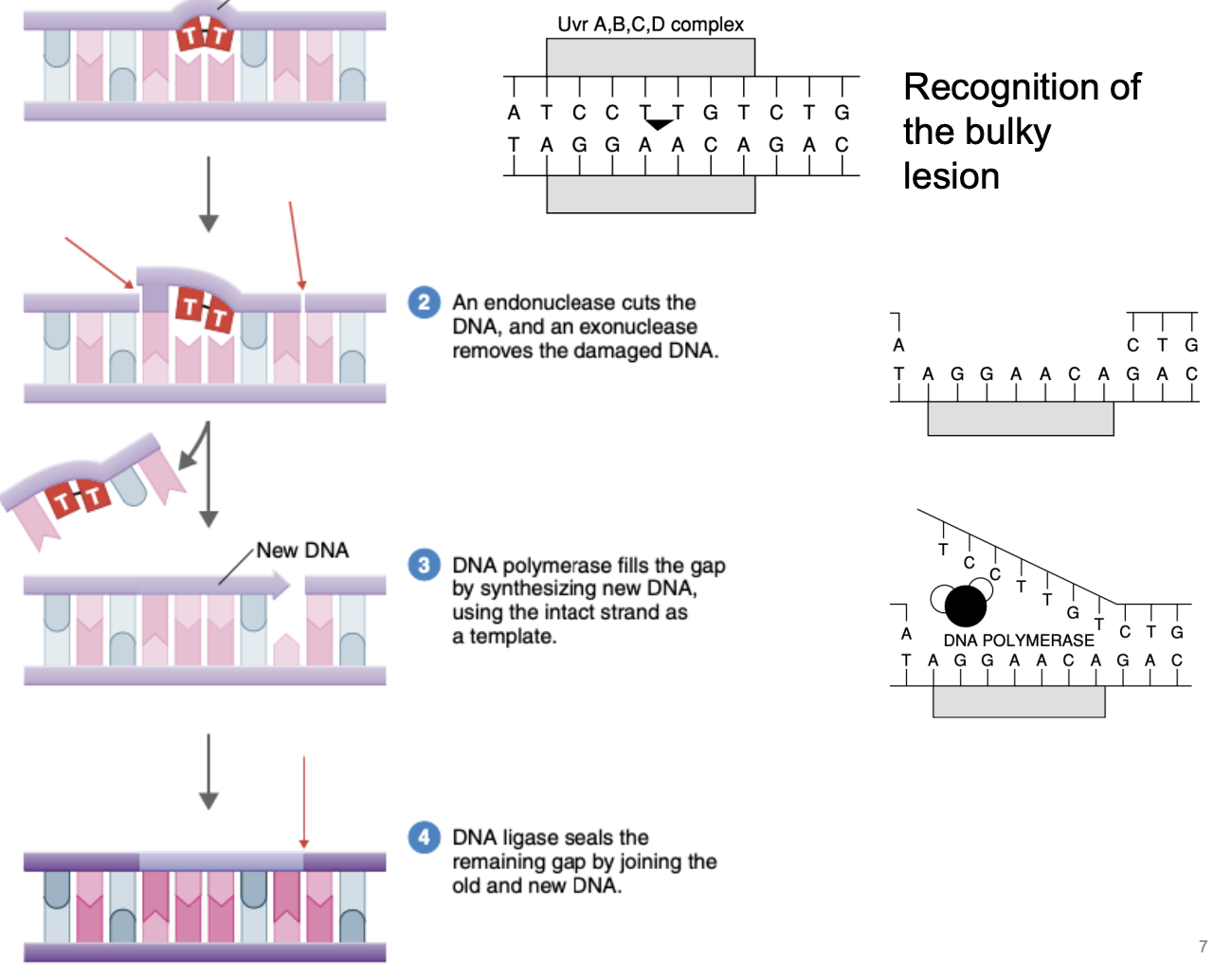
Explain nucleotide excision repair
Recognition of bulky lesion
Endonuclease cuts DNA in the middle, exonuclease removes damaged DNA
DNA polymerase fills the gap by synthesizing new DNA, using intact complementary strand as template
DNA ligase seals remaining gap by joining old & new DNA
Repairs bulky lesions, base alterations that are results of metabolic factors (e.g., oxygen radicals)
Damaged or incorrect bases is excised as free bases creating abasic sites, which could be:
Apurinic site: lacking a purine base (A,G)
Apyrimidinic site: lacking a pyrimidine abse (C,T)
Base excision repair
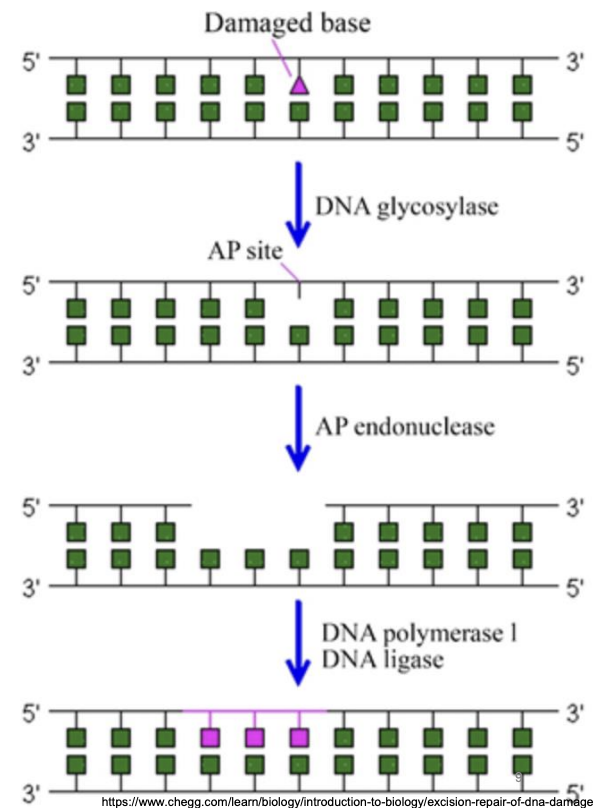
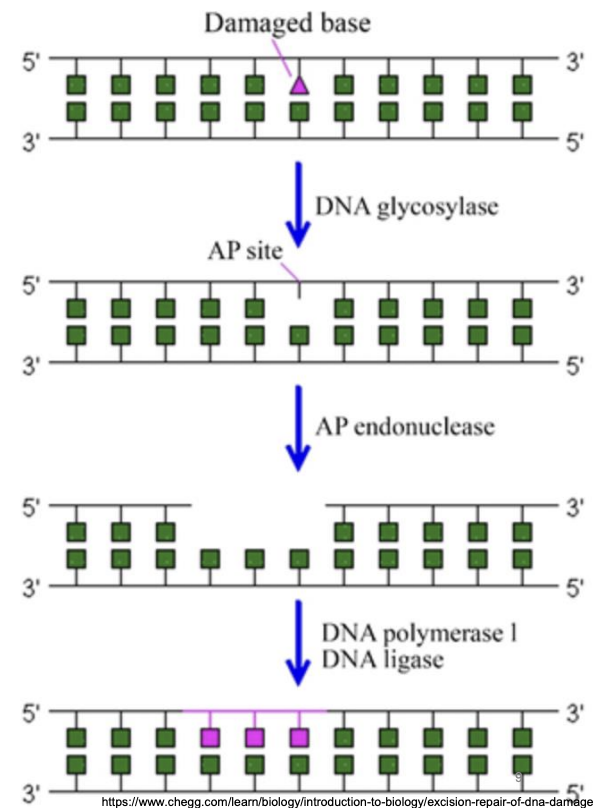
Explain base excision repair
DNA glycosylase: Hydrolysis of N-glycosidic bond between incorrect base and sugar in DNA backbone, forming AP site
AP endonuclease: Removal of AP site, excision of lesion from its 3’ side
DNA polymerase I: Filling in the resultant gap
DNA ligase: seals nicked DNA
Activated by stalled replication or major DNA damage
SOS repair system
In BER, why does excision of lesion occur from 3’ side?
AP endonuclease excises AP site from the 3’ end to facilitate the subsequent gap-filling function of DNA polymerase, which always synthesizes new DNA in 5’ to 3’ direction and recognizes 3’ OH, thus adding nucleotides from 3’ end of growing strand.
What happens to the excised bases in base excision repair?
They are released as free bases and can be reused for other anabolic processes.
Methyl-directed post-replication repair system
Repairs replicative errors within newly synthesized DNA
Allows for localized excision and resynthesis of nucleotides at site of mismatch
Mismatch excision repair
An error-prone repair system because it occurs without template
SOS repair system
This is how excision repair enzymes “know” which strand is incorrect if DNA is not physically distorted
Methylase (used for mismatch excision repair)
T/F: Mismatch excision repair is not reflected frequently because they can check if DNA is correct or not.
T/F: Mobile genetic elements are essential to cell
FALSE
Not essential
Mismatch excision repairs often repairs what type of mutation?
Spontaneous mutations
Adds a methyl group to select bases soon after DNA strand is made
Methylase
Segments of DNA with genes encoding their own replication, recombination, transmission
Mobile genetic elements (MGEs)
Who discovered methylase?
Hamilton Smith (1970)
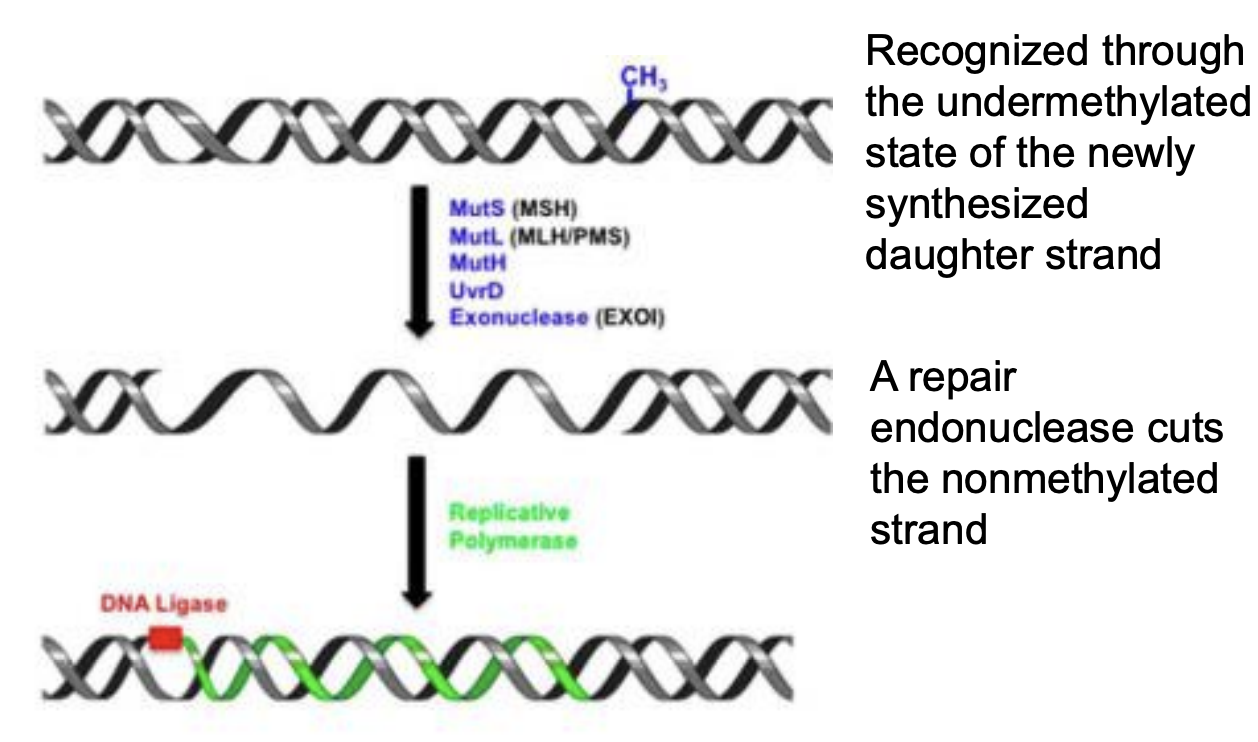
Explain mismatch excision repair
MMR proteins, such as MutS (MSH), scans and recognizes mismatched base pairs in newly synthesized DNA strand
Newly synthesized DNA strand is identified by its undermethylated state compared to parental DNA, which is already methylated
Repair endonuclease (MutH) cuts unmethylated daughter strand near mismatch site
Exonuclease removes portion of strand including mismatched base
DNA polymerase fills gap
DNA ligase seals final nick
Segments of DNA that have genes encoding their own replication, recombination, transmission rpct
Not essential to cell
Mobile genetic elements
SOS repair system is activated by _
Stalled replication or major DNA damage
Activated by stalled replication or major DNA damage
Allows DNA repair to occur without template; hence, it’s an error-prone repair
Mutations induced by this are better than cell death
SOS Repair System
SOS repair requires activation of _
RecA protein
Explain SOS repair system response
RecA protein partially repressed by LexA protein
DNA damage activates RecA, in turn, activating LexA protease activity
LexA becomes degraded
SOS response genes it represses become derepressed
dinB: encodes error-prone DNA polymerase IV
uvrA: encodes uvrA protein involved in error-free DNA repair
umuCD: encodes umuCD protein involved in error-prone repair
lexA: partially represses recA
SOS repair gene that partially represses RecA
LexA
SOS repair gene that encodes protein involved in error-free DNA repair
uvrA
SOS repair gene that encodes protein involved in error-prone DNA repair
umuCD
SOS repair gene that encodes error-prone DNA polymerase IV
dinB
Lateral gene transfer
Movement of genetic material between bacteria OTHER THAN by descent
Horizontal gene transfer
Sum of all the mobile genetic elements in a genome; not essential
Mobilome
T/F: Horizontal gene transfer involves genome replication but no cell division, unlike VGT which involves both.
TRUE
VGT = Genome replication, cell division
HGT = Genome replication
Transfer of genetic material by descent, i.e., from parent to offspring
Vertical gene transfer
Significance of HGT
Allows cells to quickly acquire new characteristics (e.g., metabolic diversity)
Provide source of nutrients (e.g., carbon, nitrogen, phosphorus)
Facilitates horizontal gene transfer
Mobilome
Facilitates HGT
Sum of all mobile genetic elements in a genome
Mobilome
5 types of mobile genetic elements
ppiti
Plasmid
Prophage
Insertion sequence
Transposon
Integron
Extrachromosomal circular dsDNA
Genes (transferred) are not necessary for growth
Plasmid
Homologous vs. Nonhomologous recombination
Homologous: requires similar sequences to integrate genetic material
Nonhomologous: integrates genetic material into random locations in a genome
Identical sequences oriented in opposite directions
Inverted repeats
Catalyzes nonhomologous recombination
Transposase
Integrated viral genomes
e.g., Lysogenic bacteriophage
Prophage
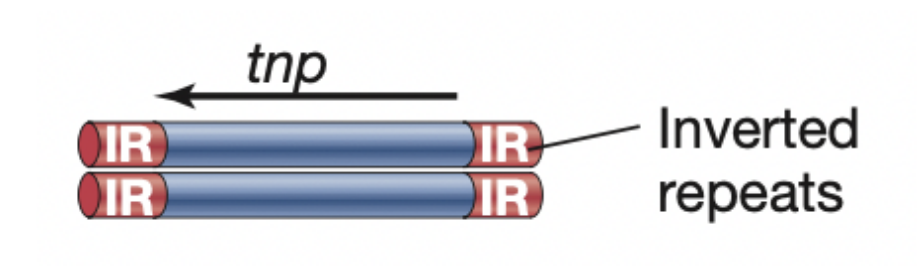
Which MGE
Insertion sequence
Facilitates movement of transposable elements; recognizes and binds to inverted repeats
Transposase

Which MGE
Transposon
Can integrate viral DNA into host’s genome
Lysogenic bacteriophage
Larger than insertion sequence but similar in function
Contains multiple genes (conjugation and other functions)
Transposon
Transposons moves between different host DNA molecules through _
Transposase
2 types of transposition
Conservative transposition
Transposon is excised from donor
Product
Donor DNA with break
Transposon in new location
Constant number of transposons bc donor loses transposition genes
Replicative transposition
Transposon is replicated
Product
Undamaged donor DNA
Transposon in new location
Increase in transposons
Simple mobile elements with tranposase gene (coding for transposase crucial to its mobility, cutting & inserting sequence from 1 location to another)
Flanked by inverted repeats (recognized by transposase, which catalyzes nonhomologous recombination)
Insertion sequence
T/F: Conservative transposition maintains a constant number of transposable elements in the genome, as the original element is removed when the new copy is inserted.
TRUE
Protein-coding genes
DNA sequences with potential to be translated into protein
Open reading frame (ORF)
Often found in other mobile elements (e.g., plasmids, transposons)
Integron
Identical sequences oriented in same direction
Direct repeats
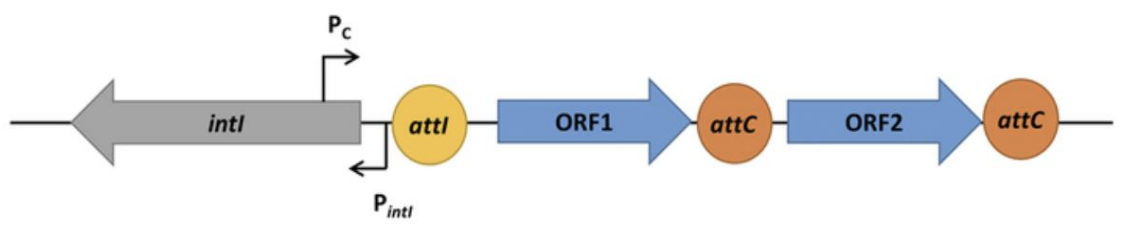
4 components of integron, explain image
Integrase (intl gene): catalyzes site-specific recombination of DNA at integron recombination site
Gene cassette: consists of 1 or more genes flanked by a series of recombination sites
Promoter (Pc): drives expression of genes within cassette
*Pintl = promoter for integrase gene
Recombination site (attI, attC): contains direct repeats
*ORF1, ORF2 = open reading frames (DNA sequences with potential to be translated into protein; protein-coding genes)
T/F: Inserting a transposon into a gene can render it nonfunctional.
TRUE
Inserting transposon into a gene disrupts its coding sequence or regulatory elements, potentially rendering it nonfunctional. This gene inactivation method is actually used by researchers to study gene function and the effects of gene inactivation on the organism.
Mechanisms of HGT
Transformation
Transduction
Conjugation
Membrane vesicle-mediated HGT
Cells that are able to take up DNA and be transformed
Competent cell
How to make cells competent in the lab (or increase their efficiency)
Since DNA is negatively charged, cells can be made more positive through alteration of its surface properties using NaCl or sucrose, for instance, to make it more efficient at attracting DNA.
Naked DNA could either be _
Cells free
DNA fragment from medium
Uptake by competent cell of naked DNA (cells free or DNA fragment from medium)
Transformation
Refers to degree by which competent cell can readily and efficiently uptake DNA
Efficiency
High-efficiency bacteria
Bacillus
Poor-efficiency bacteria
Escherichia coli
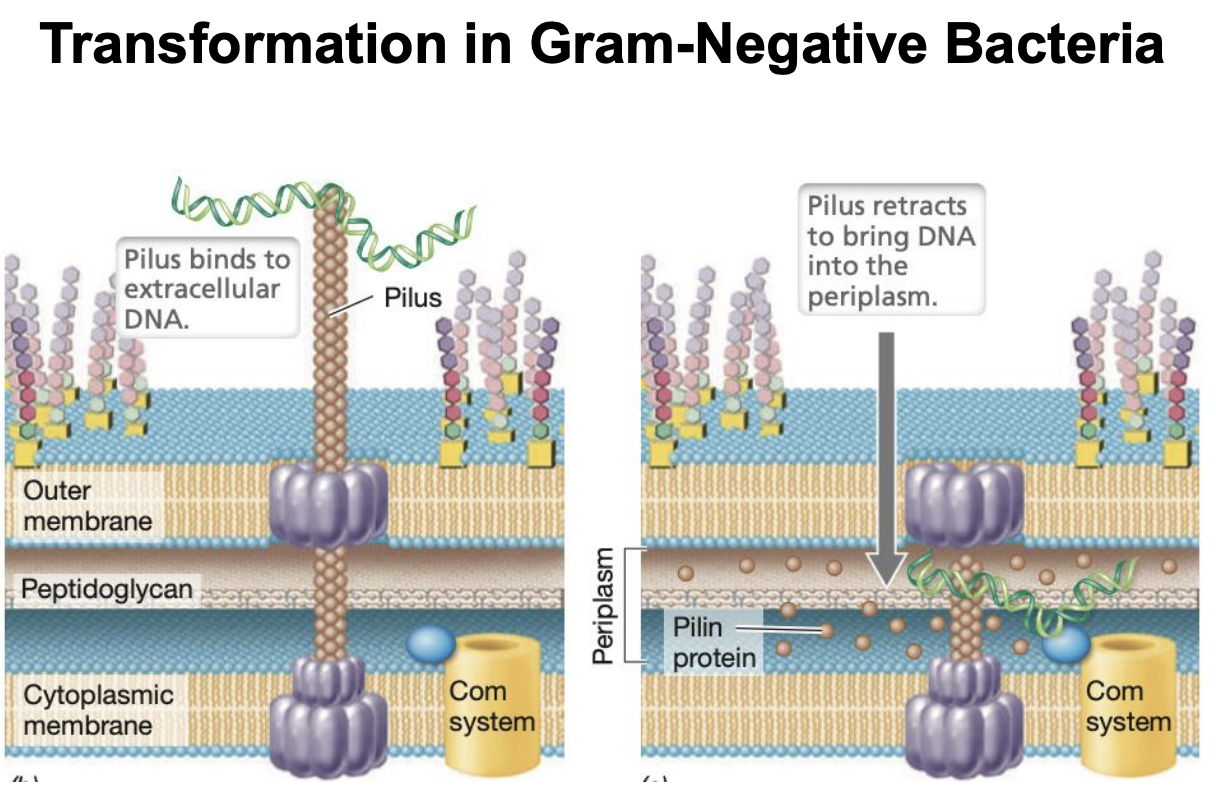
Explain transformation in Gram (-) bacteria
Pilus extending from IM to extracellular environment to bind to extracellular DNA
Pilus retracting to bring in DNA into periplasm
*Type IV pili
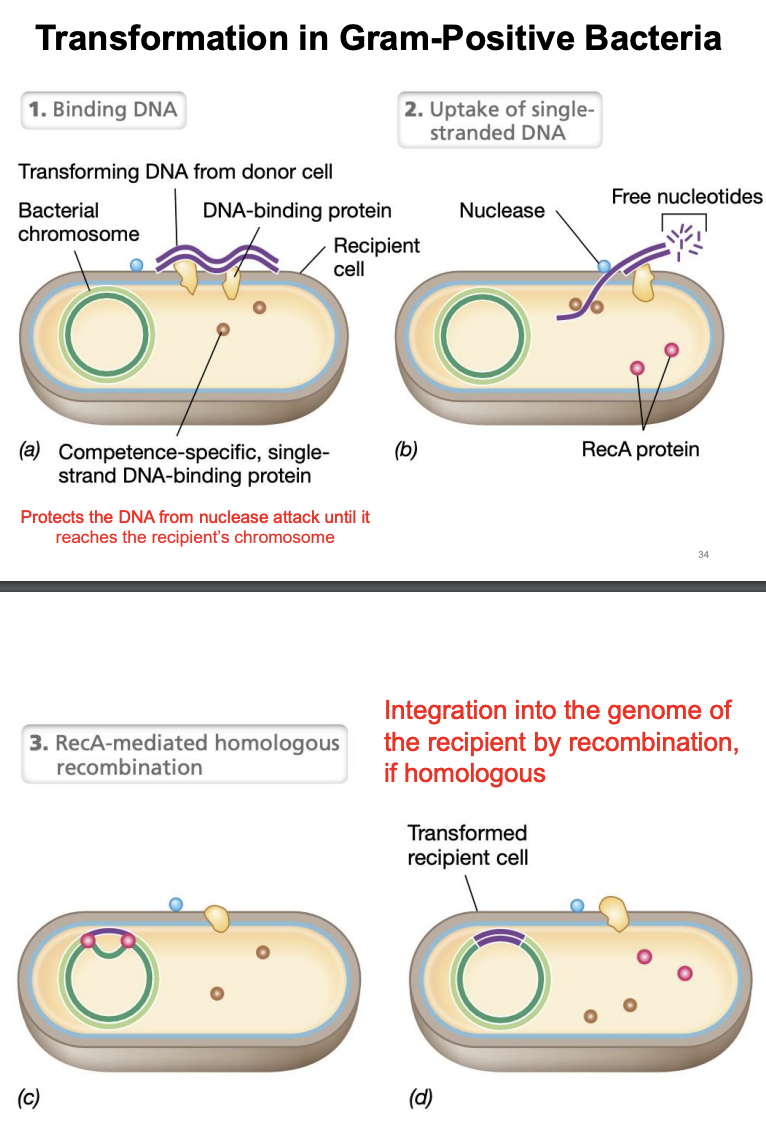
Explain transformation in Gram (+) bacteria
Binding of DNA
Recipient cell binds to naked DNA through help of DNA-binding proteins on its surface
Uptake of single-stranded DNA
Nucleases break down one of strands of dsDNA
Remaining strand is taken into recipient cell while being protected by single-stranded DNA binding proteins
RecA-mediated homologous recombination
If incoming DNA is homologous, RecA protein facilitates its integration into recipient chromosome via homologous recombination
During transformation in gram(+), what protects the DNA from nuclease attack until it reaches the recipient’s chromosome?
Single-stranded DNA binding proteins (ssDBPs)
Transfer of genetic material via direct cell-to-cell contact (mating)
Conjugation
T/F: Conjugation is a plasmid-encoded mechanism
TRUE
What structure facilitates conjugation?
Conjugation pilus
T/F: Conjugation can mediate DNA transfer between closely related cells only.
FALSE
Conjugation can mediate DNA transfer between closely related cells or between more distantly related cells
Unidirectional transfer of genetic material
Conjugation
Give example of conjugation, explain
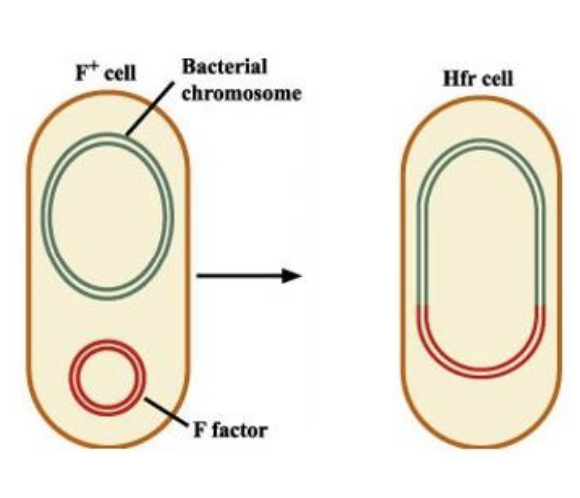
Fertility plasmid transferred via conjugation (unidirectional transfer for F+, F-)
Donor strain = F+, Hfr
F+ = nonintegrated F plasmid
Hfr = F plasmid integrated into chromosome
High frequency of recombination
Recipient = F-
T/F: If donor cell is F+, its recipient will become a donor. If donor cell is Hfr, its recipient remains a recipient.
TRUE
The entire F plasmid is sometimes too large to be fully transferred and is thus cut off in the case of Hfr recipient.
T/F: The recipient becomes Hfr only if the entire F plasmid, including the chromosomal region where it is integrated, is transferred
TRUE
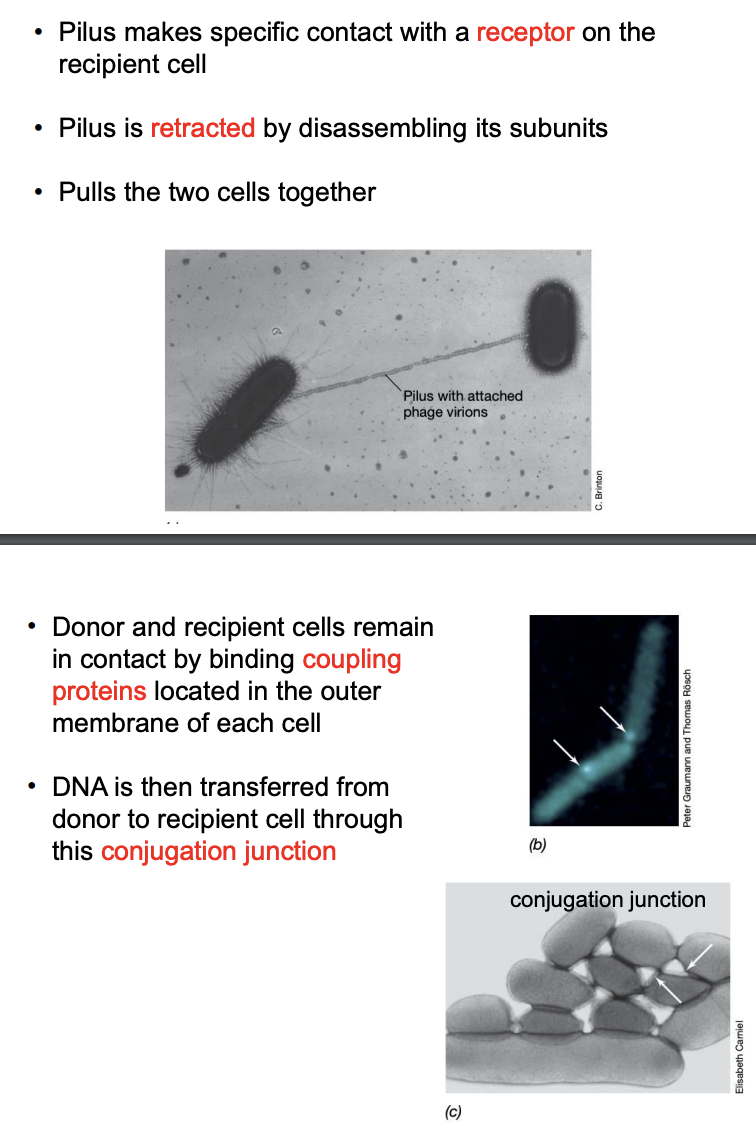
Explain general process of conjugation
Pilus of donor cell makes specific contact with receptor on recipient cell
Pilus retracts by disassembling its units, pulling 2 cells together
Donor and recipient remains in contact by binding coupling proteins in outer membrane of each cell
DNA is transferred from donor to recipient through conjugation junction
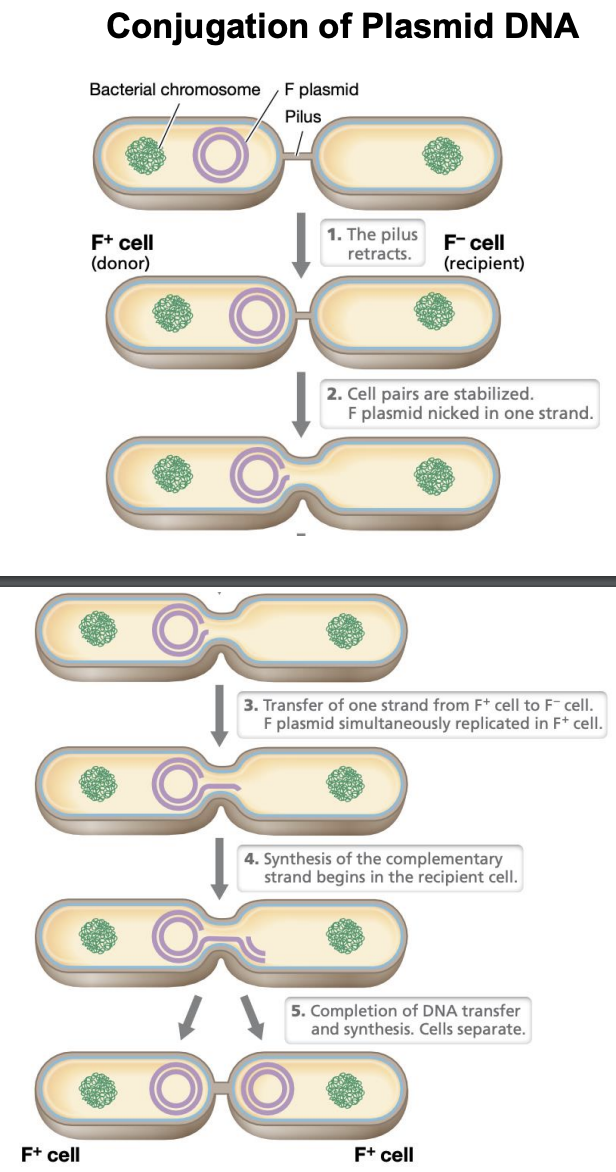
Explain conjugation of PLASMID DNA
Pilus retracts
Cells are stabilized; F plasmid nicked in 1 strand
Transfer of 1 strand from F+ to F-
Simultaneous replication of F plasmid in F+
Synthesis of complementary strand begins in recipient cell
Complete DNA transfer & synthesis. Cells separate
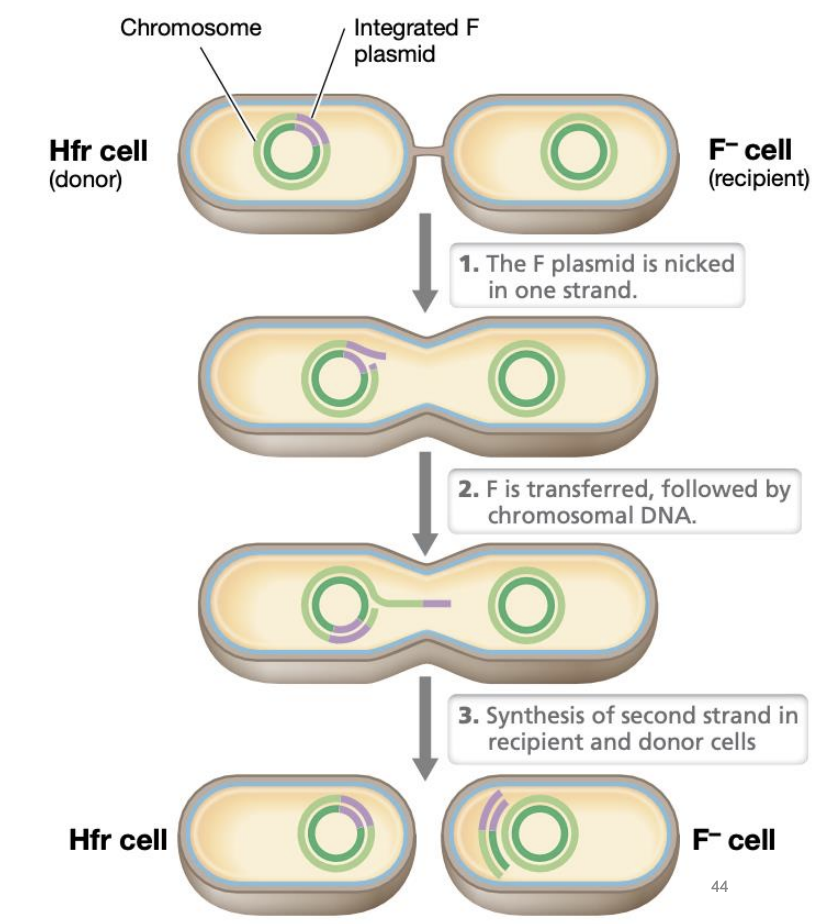
Explain transfer of CHROMOSOMAL DNA via conjugation
F plasmid nicked in 1 strand
F plasmid is transferred, followed by chromosomal DNA
Synthesis of second strand in both donor and recipient cells
Site where DNA transfer begins during conjugation
oriT (origin of transfer)
Gene transfer mediated by phages (transducing particles) containing bacterial DNA (donor genes)
Does not require cell-to-cell contact
Unidirectional (from phage to transductant)
Transduction
Host genes are occasionally and randomly packaged in phage head
Lytic phage
Low-frequency transfer: bc only a few phage particles will contain bacterial DNA
Generalized transduction
Transfers very specific genes
Occurs only with temperate and lysogenic viruses
Extremely efficient and selective transfer
Specialized transduction
Phage DNA integrated into host genome
Prophage
When portion of host DNA is exchanged for phage DNA
Specialized transduction
T/F: In specialized transduction, only bacterial genes near the prophage integration site are transferred because the excision process is not random.
TRUE
Why generalized transduction is of low frequency compared to specialized?
Because generalized transduction occurs randomly and thus not every phage it produces will contain the viral DNA, while specialized transduction forms lysogenic phages, all of which with viral DNA integrated.