Week 5-Pathogenesis of human retroviruses: family Retroviridae
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
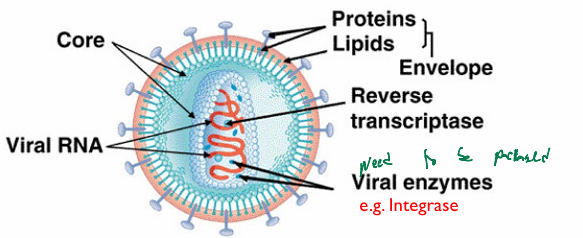
Retroviridae: virion structure
Spherical, enveloped particles
icosahedral or conical capsids
contains reverse transcriptase intrase within capsid (required; needs to be packaged)
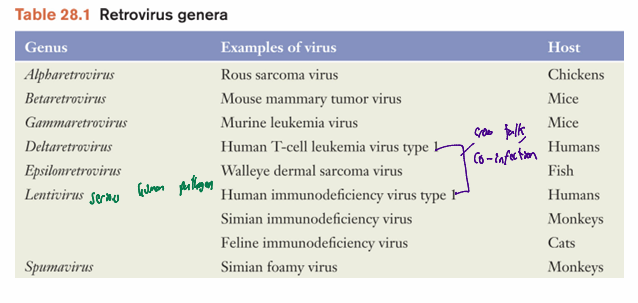
Retroviridae: classification
infect wide range of organisms
can cause AIDS and leukemia in humans
Retroviridae genome
positive- sense ss-stranded RNA (7-10Kb)
complexed with the nucleocapsid protein (NC)
2 identical copies of the (+)RNA in a dimer form are packaged into the virion
are identical sequences, not base pairs
specific cellular tRNAs bound to genome RNAs
originate from tRNA from host cells
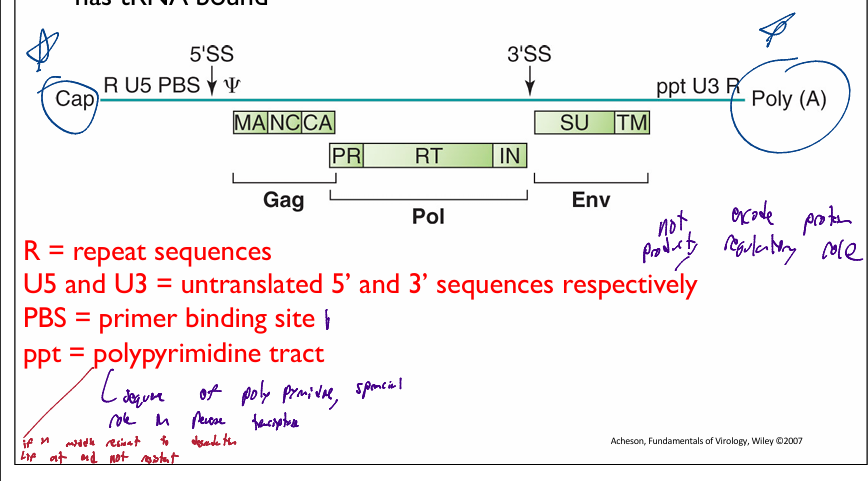
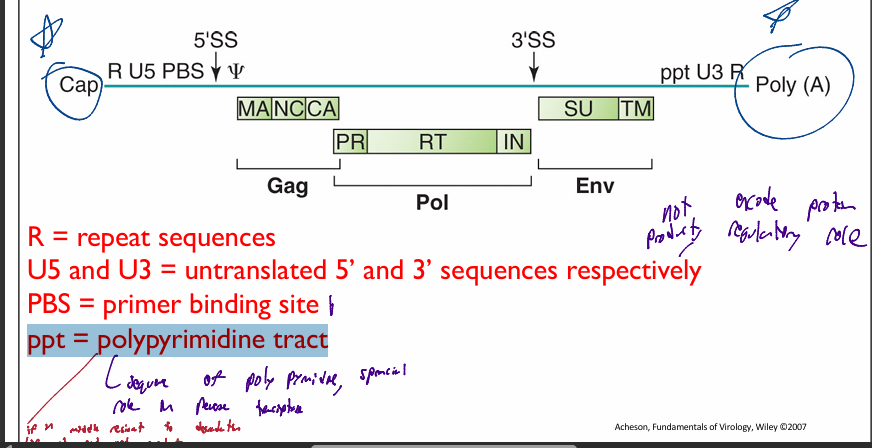
components of Retroviridae genome
R = repeat sequences
U5 and U3 = untranslated 5’ and 3’ sequences respectively
PBS = primer binding site
where tRNA binds to act as a primer for reverse transcriptase
ppt = polypyrimidine tract
if in middle of sequence can not be degraded
if att end of genomic sequence can be degraded
important during reverse transcriptase
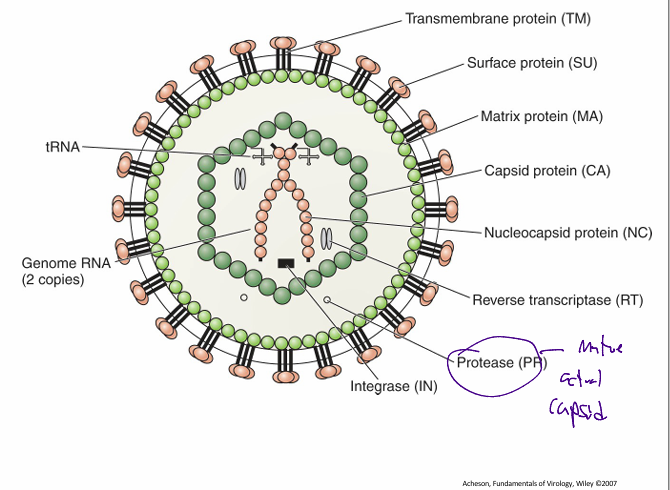
Retroviridae: virion components
envelope contains
a surface protein (SU) which is bound to the transmembrane protein (TM) that traverses the bilayer
Viral matrix protein (MA)
coats the inner side of the membrane
capsid protein (CA)
helps form the icosahedral or conical capsid
core of the virus contains 3 main virally encoded enzymes
reverse transcriptase (RT)
integrase (IN)
protease (PR)
Retroviridae: cell entry
enter cells by the fusion pathway
Viral SU protein interacts with cell surface receptors
Envelope fuses with plasma membrane or is endocytosed followed by fusion within low pH endosomes
Viral SU goes through conformational change upon receptor binding, exposing the hydrophobic region of TM protein, which inserts into the cell membrane
Early phase includes entry, making a DNA copy of its RNA genome, and integrating it into the cellular genome
overview of Retroviridae: reverse transcription
Viral RNA is converted into double-stranded DNA copy by reverse transcription
results in the production of proviral DNA
RT is a dimer
has RNA-dependent (or DNA-dependent) DNA polymerase activity
has ribonuclease H activity
RNAse H selectively destroys the RNA in an RNA-DNA hybrid
lack of proofreading capability leads to high mutation rate
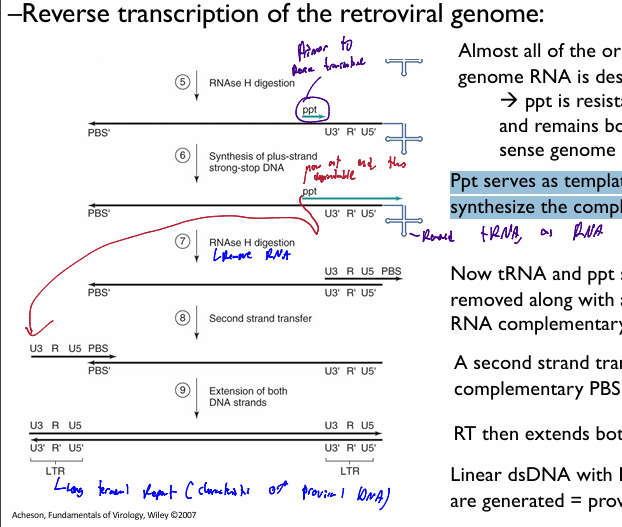
steps for Retroviridae: reverse transcription
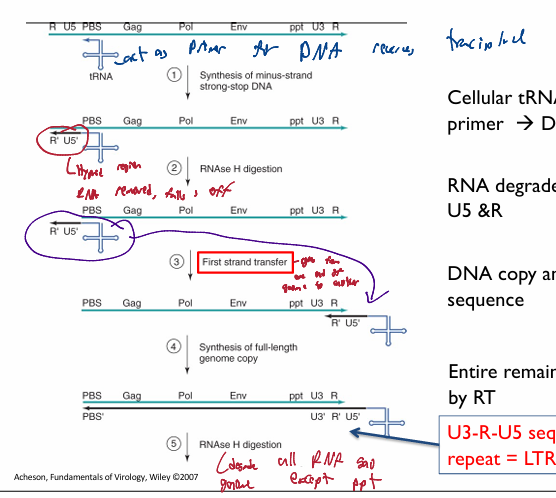
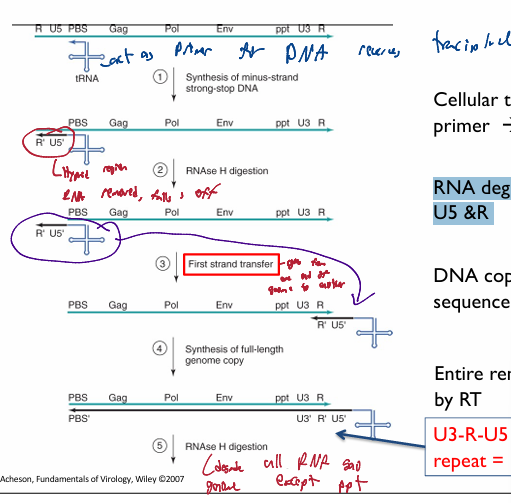
Cellular tRNA bound to PBS used as primer → DNA copy of U5 & R
makes (-) DNA strand for begining region
DNA-RNA hybrid on 5’end of original genomic DNA
RNAse H digestion targets DNA-RNA hybrid; resulting in only the RNA region being degraded → leaves DNA copy of U5 &R and tRNA primer
first strand transfer. The 5’ R section compliments te 3’ R section. Thus the (-) U5 and R DNA segementt attach to the 3’ end with the tRNA
DNA copy anneals to 3’ end R sequence
reverse transcriptase makes a full-length (-) sense DNA genome copy
resuling U3-R-U5 sequence is a long terminal repeat = LTR
a pair of identical sequences of DNA, several hundred base pairs long, flank genes/pseudogenes/viral dna. forms a retrotransposon or an endogenous retrovirus or a retroviral provirus
Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome
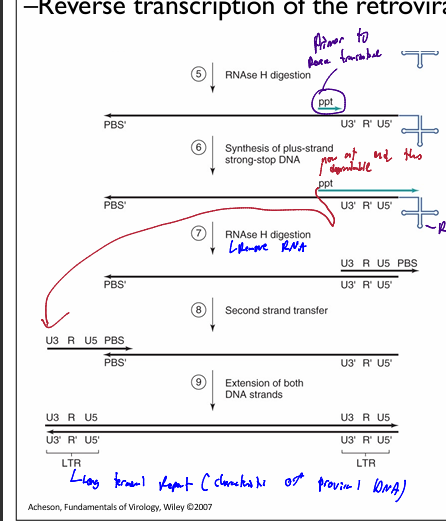
RNAse H digestion removes almost all original RNA genome template, exceptt ppt (as it was in middle of sequence
Ppt serves as template for RT to synthesize a complementary strand to tthe 3’ end of the made (-)DNA genome
results in DNA RNA hybrid at the 3’ end (-) DNA stand
RNAse digest ppt & tRNA, leaving only U3-R-U5 PBS (+) RNA genome
Second strand transfer→ the PBS (+) moves and annealls to the PBS (-)
RT then extends both DNA strands
Linear dsDNA with LTRs on both ends are generated = proviral DNA
Retroviridae: integration
A copy of proviral DNA is integrated into the cellular genome at a random site
some sites more likely to be integraded (if not methylated or compressed)
Carried out by the enzyme integrase (IN)
stably integrated into host genome
Usually have to wait for nuclear envelope to break down for integration
Retroviridae: transcription & translation
Host cell transcribes & translates virus genome
translation of the multiple mRNAs
processing enzymes cleave the polyproteins into single proteins
requires viral protease to cleave polyprotein
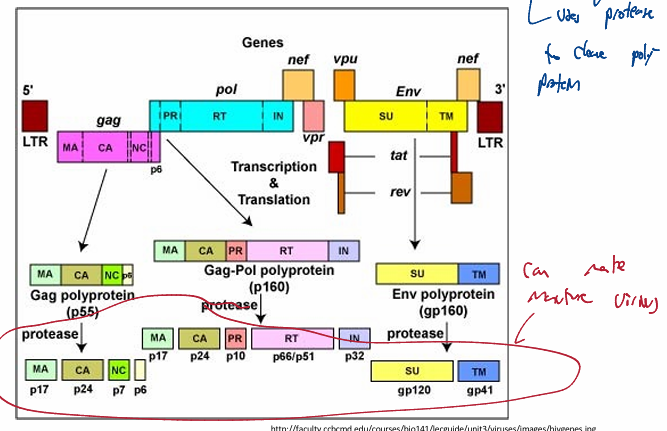
Retroviridae: budding & cell exit
Late phase involves expression of viral RNA, viral protein synthesis and assembly of virions
Envelope proteins go through the host secretory pathway= expressed on PM
budding occurs at sites where envelope proteins cluster
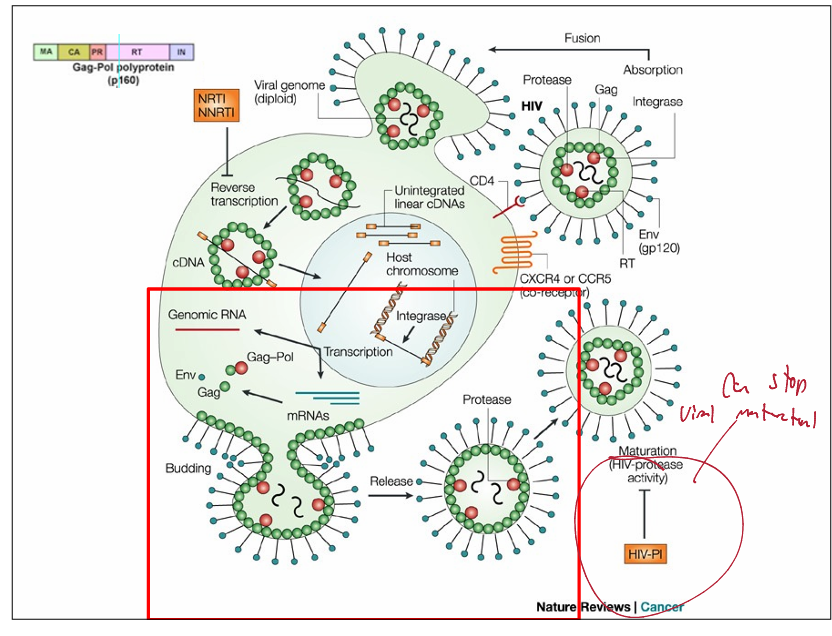
Retroviridae antiviral
NRTI & NNRTI: inhibits reverse transcription
HIV-P1: stops viral maturation
HIV buds off then comples virion maturation
Retroviridae: Lentiviruses
for slow progression of disease
results Human immunodeficiency virus types 1 and 2 (HIV-1, HIV-2)
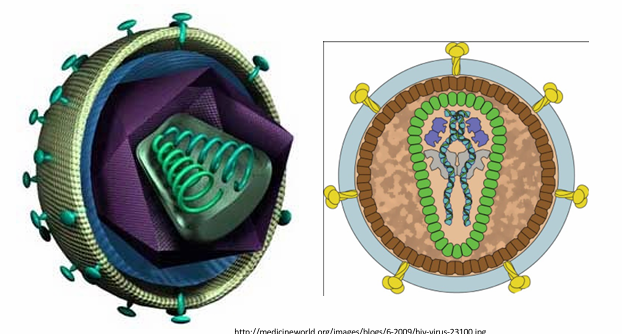
Human Immunodeficiency Virus Type I genome structure
Spherical enveloped particle
100nm
conical capsid
9.3Kb ssRNA positive sense genome (2 RNAs/virion)
Lysine tRNA bound to genome RNAs
is a complex retrovirus
Human Immunodeficiency Virus Type I genome structure
complex retrovirus (encodes 25 mRNAs)
simpler retroviruses make 2 mRNAs
Uses alternative splicing leads to several mRNAs; makes
structural proteins
regulatory proteins
Human Immunodeficiency Virus Type I encodes
three capsid proteins
matrix (MA)
capsid (CA)
nucleocapsid (NC)
Three enzymes
protease (PR)
reverse transcriptase (RT)
integrase (IN)
Two envelope proteins
surface (SU)
transmembrane (TM)
Five regulatory proteins (6 non-structural)
Vif (viral infectivity factor)
Vpu (virion protein unique to HIV-1)
Tat (transactivator of transcription)
Rev (regulator of expression of virion proteins)
Nef (negative effector)
Vpr (virion protein R)
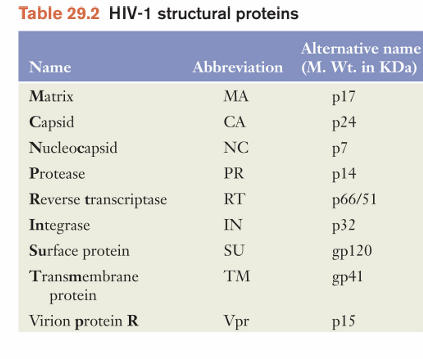
HIV-1 cell entry
targets cells of the immune system by recognizing CD4 antigens and chemokine receptors
CD4 is found on both T lymphocytes and macrophages
A co-receptor of either CCR5 or CXCR4 is also required
CXCR4 is expressed on T cells
CCR5 is expressed on macrophages
T-cell tropic or macrophage tropic virus strains
example of Model of HIV-1 entry:
gp120 interacts with CD4
conformational change exposes region on gp120 that interacts with chemokine receptor
fusion domain of gp41 now becomes exposed
close proximity of viral and cell membrane induces fusion = viral nucleocapsid enters cell cytosoll
Unlike other retroviruses, HIV-1 directs transport of proviral DNA into the cell nucleus
allows for productive infection in non-dividing cells
many regulatory proteins aid in this process
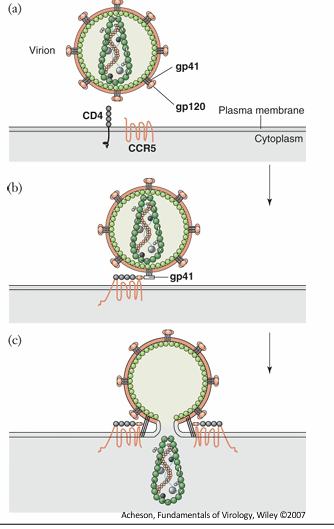
oncogenic retroviruses
Acute transforming retroviruses express mutated forms of cellular growth signaling proteins
Acute transforming retroviruses express mutated forms of cellular growth signaling proteins
growth factors, receptors, transcription factors etc.
bring in genes simillar to host, which is unregulated (like growth factor), result in rapidly dviding cells cancer
example
Viral Src is nearly identical to cellular c-src
codes for a protein tyrosine kinase
under the control of a viral promoter src is unregulated=uncontrolled cell growth = tumours
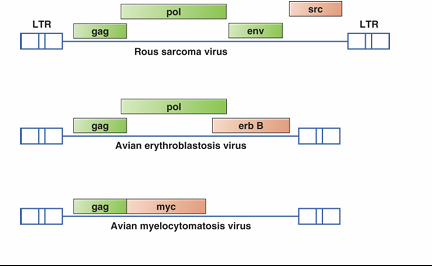
HIV & AIDS
Acquired immune deficiency syndrome (AIDS) was first described in 1981
HIV replicates in and kills lymphocytes and macrophages
due tto infectious immune cell
-Infection results in depletion of CD4+ T cells, resulting immune-incompetent
Opportunistic infections by other pathogens are often fatal
HIV is transmitted through sexual contact and blood exchange
sharing needles
still a global HIV epidemic, but reduced numbers significantly (good health promotions, trying to reduce it more)
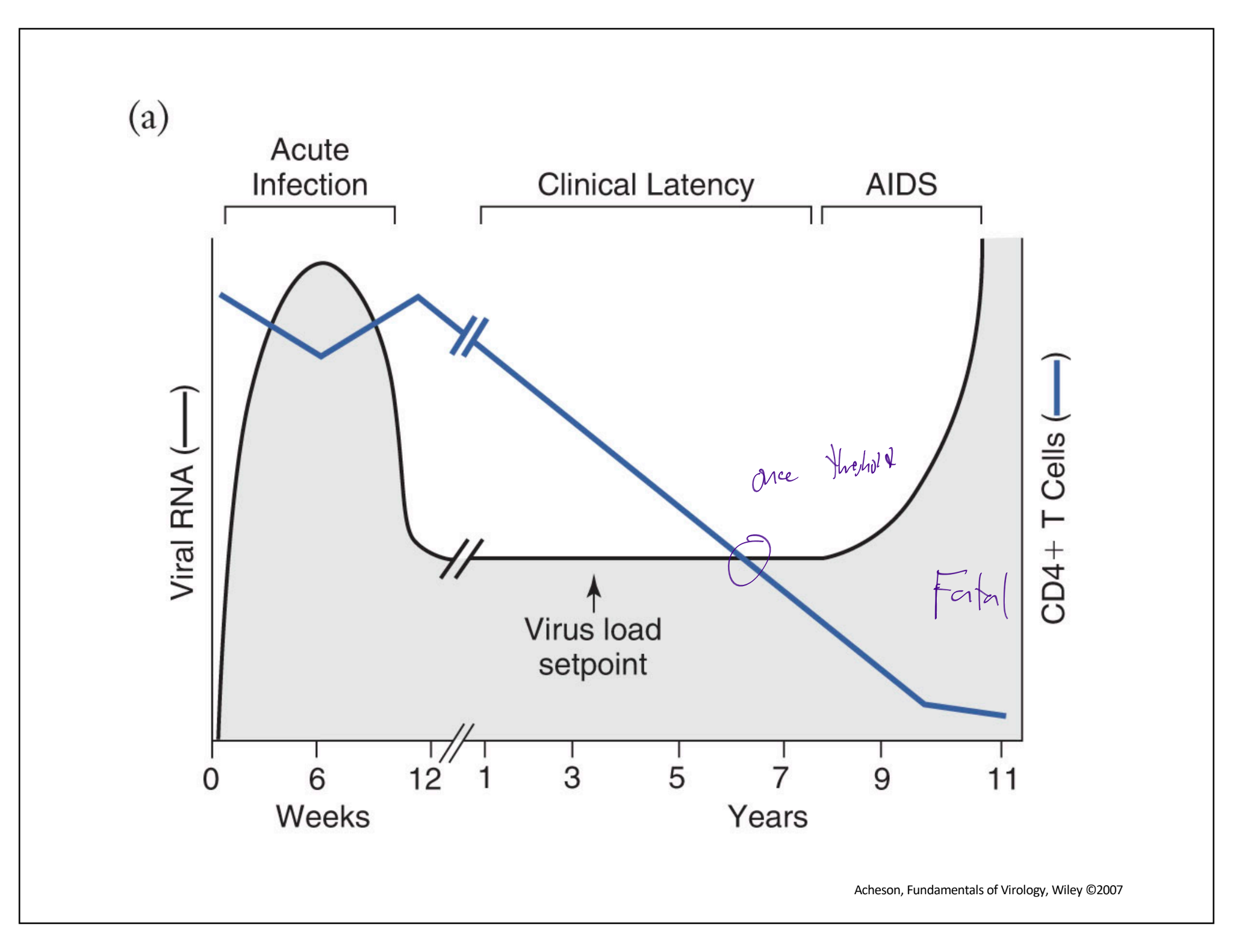
Stages of AIDS
Acute infection
inital infection of HIV virion
High viral RNA, slight CD4+ T cells
some immune response and thus flu like symtoms
spike in CD4+ cells
virions begin to infect/enter immune cells
Clinical latency
virus RNA levels consistent (virus load setpoint)
steady decline of CD4+ cells
AIDS
once CD4+ deacrease pass a threshold result in AIDS
lack of immune cells, increase viral RNA (nothing stopping them, and will multiply)
HIV & treatment
No vaccine developed
Imbokodo vaccine study:
“Mosaic” vaccine that combines epitopes from many different HIV strains, making a truly global vaccine
Enrolled women in sub-Saharan Africa – 2600 participants vaccinated
induced neutralizing antibodies in animals (but not shown in humans, thus was canceled).
Used Alum adjuvants
Antiviral drugs can control HIV-1 infection and prevent disease progression
Treatment involves a combination of drugs targeted at various steps in the viral life cycle (e.g. reverse transcription, fusion, protease processing, integration etc.) = anti-retroviral therapy
e.x
PrEP (pre-exposure prophylaxis)
for after exposure