late night 116-162 flashcards chapter 6
1/94
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
95 Terms
What is Cleland nomenclature used for?
A shorthand notation for describing reactions with multiple substrates and products.
How are substrates labeled in Cleland notation?
How are products labeled in Cleland notation?
SUB: As A, B, C, D, in the order they bind to the enzyme.
PRO: As P, Q, S, T, in the order they dissociate from the enzyme.
What do uni-, bi-, ter-, and quad- refer to in Cleland nomenclature?
The number of substrates:
Uni = 1 substrate
Bi = 2 substrates
Ter = 3 substrates
Quad = 4 substrates
How is the enzyme represented?
As E, but if modified during the reaction, successive forms are F, G, etc.
Give an example of Cleland notation for a two-substrate reaction where A binds first, B second, P is released first, and Q second.
A + B → P + Q with enzyme forms: E → F → E
What is a single displacement reaction?
Both substrates bind to the enzyme before products are formed.
In a random single displacement reaction, which step is rate-limiting?
What does "random" mean in this context?
AEB → PEQ
Substrates can bind in any order.
Give an example of an enzyme with a random, single-displacement mechanism.
Creatine Kinase
What determines the overall direction of a single-displacement reaction?
The relative concentrations of the substrates and products (e.g., ATP, ADP, Cr, CrP) and the equilibrium constant for the reaction.
T/F Most kinases have a random, single-displacement mechanism.
True
What is characteristic of product release in an ordered single-displacement reaction?
There is an ordered release of products.
Can substrate B bind to the enzyme in the absence of substrate A for orderedd single?
No, B will not bind to E unless A is already bound.
What happens in bisubstrate single-displacement reactions (both random and ordered) when substrate concentration increases?
The reaction reaches saturation, and the curve no longer shifts.
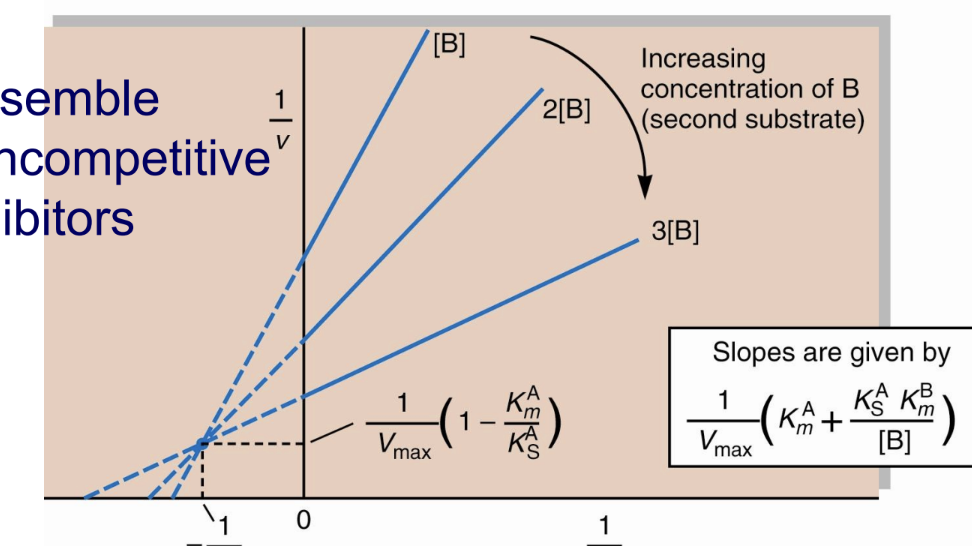
What do bisubstrate single-displacement reactions resemble in terms of inhibition?
Noncompetitive inhibitors.Vmax dec, Km stays same
What is a double displacement (ping-pong) reaction?
The two substrates bind and react separately in a ping-pong manner.
What is E’ in a double displacement reaction?
E’ is a covalently modified enzyme intermediate.
In double displacement reactions, when is the product of the first substrate released?
The product of substrate A (P) is released before the enzyme reacts with the second substrate (B).
What does “burst” kinetics indicate in a double displacement reaction?
The first product (P) appears rapidly before the second product (Q) is formed, reflecting the two-step ping-pong mechanism.
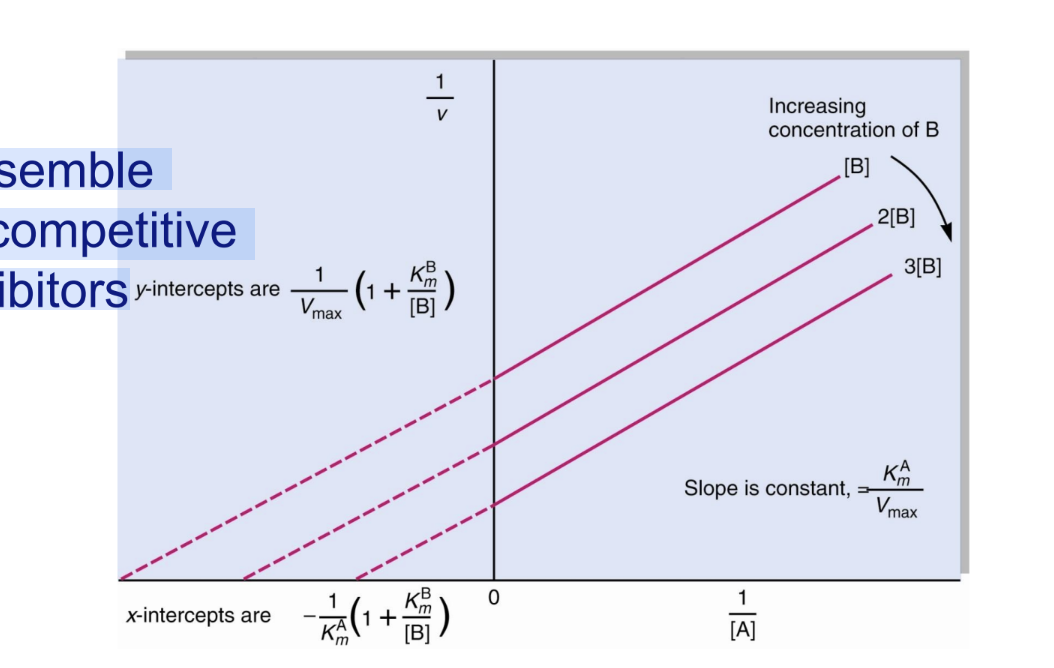
What type of inhibition do double displacement bisubstrate reactions resemble?
Uncompetitive inhibitors.
What is the Lineweaver–Burk pattern for noncompetitive inhibition?
Lines intersect on the x-axis because Km is unchanged.
What is the Lineweaver–Burk pattern for uncompetitive inhibition/double dispalcement bisubstrate?
Lines are parallel because both Km and Vmax decrease proportionally.
Give an example of an enzyme that follows a double-displacement bisubstrate mechanism.
Glutamate:aspartate aminotransferase
What is the first method for diagnosing bisubstrate reactions?
Kinetic analysis using a Lineweaver–Burk plot to distinguish single vs double displacement mechanisms.
How can substrate binding assays help diagnose bisubstrate reactions?
They determine whether substrate addition is random or ordered.
What are exchange reactions used for in diagnosing bisubstrate reactions?
To test if the reverse reaction occurs in the absence of one substrate:
Double displacement: reaction still occurs
Single displacement: reaction does not occur
What are sucrose phosphorylase and maltose phosphorylase examples of?
Enzymes that perform exchange reactions — specifically double-displacement (ping-pong) mechanisms.
Write the exchange reaction catalyzed by sucrose phosphorylase.
Sucrose + Pi ↔ glucose-1-P + fructose
Write the exchange reaction catalyzed by maltose phosphorylase.
Maltose + Pi ↔ glucose-1-P + glucose
How is the exchange reaction tested experimentally?
Mix ³²P-labeled Pi with glucose-1-P and add enzyme:
G-1-P + ³²Pi ↔ Pi + G-1-³²Pi
What is the exchange reaction tested with phosphorylases?
G-1-P + ³²Pi ↔ Pi + G-1-³²Pi
Does sucrose phosphorylase catalyze the exchange reaction? What does that indicate?
Yes, it catalyzes the exchange → Double Displacement mechanism.
T/F Only double displacement (ping-pong) enzymes can catalyze exchange reactions.
True
What is the reaction mechanism for sucrose phosphorylase?
Sucrose + E ↔ E–glucose + fructose
E–glucose + Pi ↔ E + glucose-1-P
Does maltose phosphorylase catalyze the exchange reaction? What does that indicate?
❌ No, it does not catalyze the exchange → Single Displacement mechanism.
What is the reaction mechanism for maltose phosphorylase?
Maltose + E + Pi ↔ Maltose:E:Pi
Maltose:E:Pi ↔ glucose-1-P + glucose
What are serine proteases?
Enzymes that use a serine residue in their active site to catalyze peptide bond cleavage.
Name three digestive serine proteases and their form before activation.
Trypsin, chymotrypsin, and elastase — secreted as proenzymes (zymogens).
What is the function of thrombin?
A serine protease in the blood clotting cascade.
What is subtilisin?
A bacterial serine protease.
What is the function of plasmin?
Cleaves fibrin polymers in blood clots.
What does tissue plasminogen activator (tPA) do?
Cleaves the plasmin proenzyme (plasminogen) to form plasmin — used clinically to prevent heart attacks.
Why is acetylcholinesterase grouped with serine proteases?
It is not a protease, but is mechanistically similar, using a serine residue to break down acetylcholine.
What are trypsin, chymotrypsin, and elastase secreted as?
Proenzymes (zymogens).
What is the function of trypsin, chymotrypsin, and elastase?
They cleave polypeptide chains (digestive serine proteases).
How are trypsin, chymotrypsin, and elastase related?
They have similar amino acid sequences, structures, and catalytic mechanisms.
In structural diagrams of these enzymes, what do the colored dots represent?
In structural diagrams, what do the gold lines represent?
color: Amino acids that are identical across all three enzymes.
gold: Disulfide bonds that stabilize the protein structure.
What does the chymotrypsin–eglin C structure show?
It shows the catalytic triad — His57, Asp102, and Ser195 — with Ser195 positioned close to the peptide bond of the bound substrate, ready for cleavage.
How do trypsin, chymotrypsin, and elastase differ in substrate specificity?
Trypsin: Cuts after Arg or Lys (basic), except when followed by Proline
Chymotrypsin: Cuts after Phe or Tyr (aromatic)
Elastase: Cuts after small, neutral residues
If an enzyme cleaves a protein at 4 sites, how many peptide fragments are produced?
5 peptides — the number of fragments = number of cuts + 1
What are the products of a trypsin digestion?
A mixture of peptide fragments ending with Arg or Lys and one peptide from the C-terminal end of the original polypeptide.
What is trypsin’s binding pocket like and why?
Negatively charged → binds basic residues (Arg, Lys).
What is chymotrypsin’s binding pocket like and why?
Hydrophobic → binds aromatic residues (Phe, Tyr).
What is elastase’s binding pocket like and why?
Shallow, blocked by bulky residues (Thr, Val) → binds small residues.
What is the substrate-binding pocket in chymotrypsin?
Ser 189 is not the active site serine; the pocket determines substrate specificity, separate from the catalytic Ser195.
How is chymotrypsin activity assayed and what kinetic behavior is observed?
Uses an artificial substrate that produces nitrophenolate, absorbing at 400 nm
Shows burst kinetics: a fast first step followed by a slower second step
What key steps are involved in the chymotrypsin mechanism?
Acylation and deacylation of a serine residue.
What is a protease? Give an example.
An enzyme that catalyzes hydrolytic cleavage of peptide bonds; example: bovine pancreatic chymotrypsin.
Why do serine proteases display burst kinetics?
Because they form an acyl-enzyme intermediate:
Fast step: substrate cleavage forming acyl-enzyme
Slow step: attack of water to release P1 (C-terminal) and P2 (N-terminal) peptide fragments
What types of catalysis are involved in the serine protease mechanism?
A mixture of covalent catalysis and general acid–base catalysis.
What is the role of Asp102?
Orients His57 and forms a low-barrier hydrogen bond (LBHB); helps His57 act as a general acid/base.
What is the role of His57?
Acts as a general acid and base during catalysis.
What is the role of Ser195?
Forms a covalent bond with the peptide to be cleaved, turning the trigonal C into a tetrahedral intermediate.
How is the tetrahedral oxyanion intermediate stabilized?
By the backbone N-H groups of Gly193 and Ser195 (the oxyanion hole).
Summarize the catalytic cycle of serine proteases.
Substrate binds in the active site.
His57 acts as a general base, forming the E–Ser195–S covalent intermediate.
His57 is stabilized by a low-barrier hydrogen bond (LBHB).
Tetrahedral intermediate collapses, releasing the first product (P).
Stable acyl-enzyme intermediate allows water to enter.
His57 activates water for a nucleophilic attack.
Second tetrahedral intermediate collapses, releasing the second product.
Active site restored for another catalytic cycle.
How does chymotrypsin stabilize its transition states?
The two tetrahedral oxyanion transition states are stabilized by amide groups in the “oxyanion hole.”
How is the negative charge on the tetrahedral oxyanion stabilized in chymotrypsin?
By hydrogen bonding with the backbone amide (NH) groups of Ser195 and Gly193 in the oxyanion hole.
At what pH does chymotrypsin show optimal activity, and what are the states of key residues?
pH 8 → His57 is unprotonated and Ile16 is protonated.
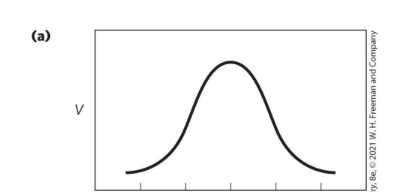
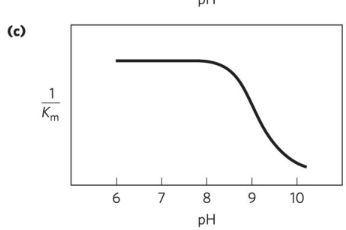
What causes the activity transition just above pH 7 for Chymotrypsin reactions?
Changes in kcat due to protonation of His57.
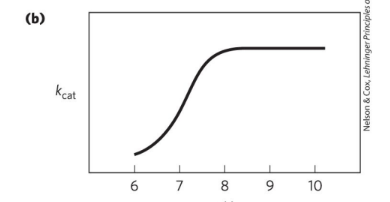
What causes the transition above pH 8.5 for Chymotrypsin reactions??
Changes in 1/Km due to ionization of the α-amino group of Ile16.
Where are catalytic triads found?
In several hydrolases and transferase enzymes, arising via divergent or convergent evolution.
What is notable about the residues in catalytic triads?
They are often far apart in the primary sequence but come together in the 3D structure.
What are the three functional residues in a catalytic triad?
Acid – orients and stabilizes the base (Asp, Glu, His)
Base – polarizes the nucleophile (His, rarely Lys)
Nucleophile – attacks the substrate (Ser, Cys, sometimes Thr)
How do aspartic proteases differ from serine proteases?
They have a different structure and mechanism and do not use covalent catalysis.
What residues are found in the active site of aspartic proteases?
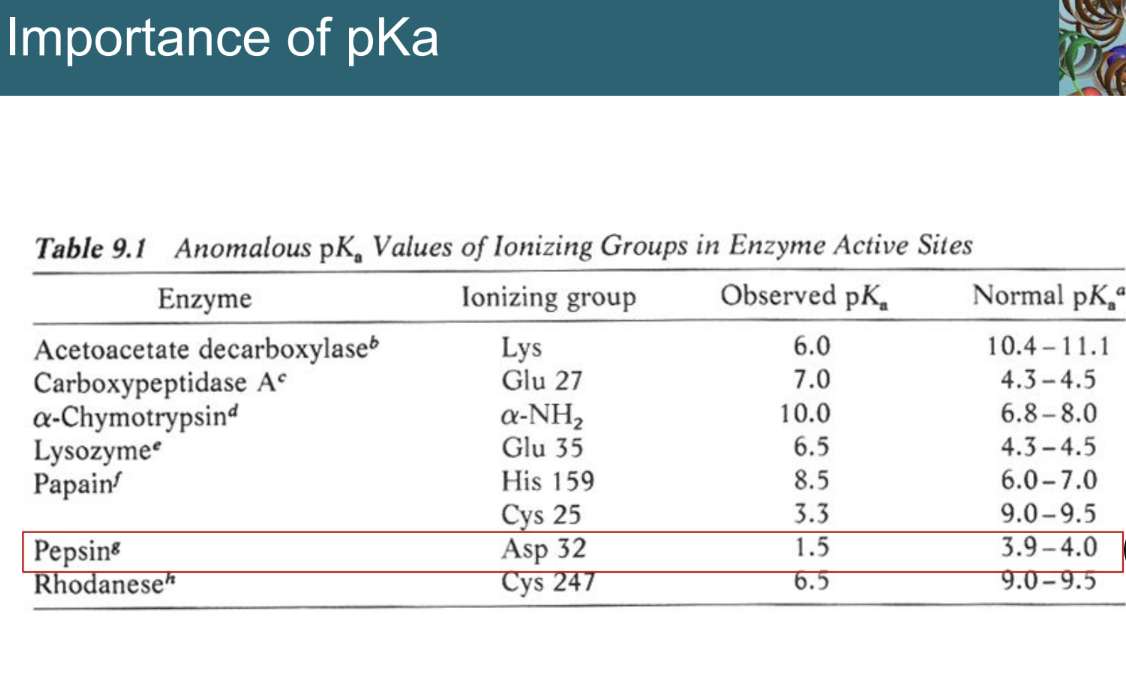
Two aspartic acids (e.g., pepsin residues 32 and 215).
What type of peptide bonds do aspartic proteases cleave?
Peptide bonds between two hydrophobic amino acids.
Do aspartic proteases use covalent catalysis?
No, they use acid-base catalysis instead.
Describe the structures of HIV-1 protease and pepsin.
HIV-1 protease: homodimer
Pepsin: monomer, N-terminal half (red), C-terminal half (blue)
In both, each lobe contributes one catalytic Asp to the active site.
What is the pH dependence of aspartic proteases?
Active at acidic pH
Mechanism requires one Asp protonated and the other Asp deprotonated when the substrate binds.
Why is the pKa of active site residues in pepsin important?
The observed pKa of the catalytic Asp residues is shifted from their normal values, allowing one to be protonated and the other deprotonated at acidic pH, which is essential for catalysis.
***JUST KNOW THAT PKA CHANGES AT ACTIVE SITE
What features are important in the mechanism of aspartic proteases?
Low-barrier hydrogen bonds (LBHBs) stabilize states E, ES, ET′, EQ′, EP′Q
Catalytic water participates in peptide bond cleavage
LBHBs allow hydrogen tunneling (electron/proton transfer)
Formation of a tetrahedral intermediate during catalysis
What is the function of HIV-1 protease?
Cleaves the polyprotein products of the HIV genome into functional viral proteins.
How does HIV-1 protease compare to mammalian aspartic proteases?
It is a remarkable mimic, functioning similarly despite being viral.
What is notable about the structure of HIV-1 protease?
It is a homodimer, which is genetically economical for the virus.
What is special about the active site of HIV-1 protease?
The active site is two-fold symmetric, with different flaps that control substrate access.
Why are protease inhibitors important for AIDS treatment?
Inhibiting HIV-1 protease prevents formation of new viral particles.
How were HIV-1 protease inhibitors developed?
Structure-based drug design has developed several inhibitors that work in the dish.
How do protease inhibitors block HIV-1 protease?
The inhibitor (e.g., Crixivan, red) binds the active site, which is covered by flaps (green), and interacts with the catalytic Asp residues (violet).
What is the purpose of pre–steady state kinetics?
To measure rate constants of individual steps in a reaction
How are pre–steady state kinetics experiments performed?
Using a stopped-flow device that allows mixing and sampling on a second or millisecond time scale.
How can pre–steady state kinetics identify the rate-limiting step?
Observation of a burst indicates a rate-limiting step (e.g., product release or enzyme conformational change) occurs after formation of the monitored product.
How does understanding enzyme mechanisms help produce antibiotics?
Peptidoglycan, a major component of bacterial cell walls, is built from polysaccharides and peptides cross-linked in steps including a transpeptidase reaction.
How do β-lactam antibiotics inhibit transpeptidase?
hey bind to the active site, forming a covalent complex that irreversibly inactivates the enzyme, blocking cell wall synthesis.
What are β-lactamases?
Enzymes that cleave β-lactam antibiotics.
What happens to bacteria that express β-lactamases?
They become resistant to β-lactam antibiotics.
How can β-lactamases be inactivated?
Using compounds that mimic the structure of a β-lactam antibiotic.