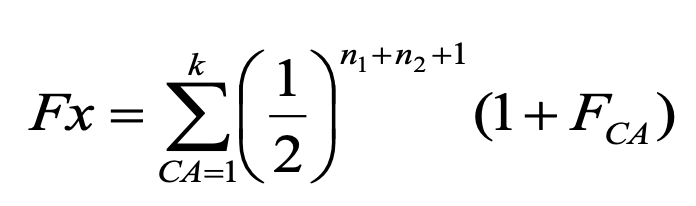Applied Genetics 2: Measures of genetic diversity
1/56
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
57 Terms
Variation
occurs at several level in the genome, decreasing in scale from chromosomal anomalies to SNPs
Diversity is only a meaningful concept where there is
Measurable variation in the genome
Population defintion
a group of individuals living in sufficiently close proximity that any member of the group could potentially reproduce with any other member.
Microsatellite
repeating sequences of 2-5 base pairs with variable numbers of repeat units
also known as short tandem repeats (STR) or simple sequence repeats (SSR) or variable tandem repeats (VNTR)
likely involved in variable gene expression
What is a DNA alignment?
Visual way to compare the same portion of DNA from multiple individuals
Where are microsatellites located?
Introns of genes
Single nucleotide polymorphisms (SNP)
locations in genome where a measurable proportion (~2-5% minimum) of the population has one nucleotide and the remainder of the population has another nucleotide
Some SNPs have
functional consequences via non-synonymous subsititution or modification of gene expression, while other don’t appear to have direct consequences
How are SNPs detected?
by sequencing but mostly species-specific SNP chips labeled with DNA probes
Amitraz-resistance SNPs Cause
A change in amino acids from isoleucine to phenylalanine which prevents the binding of amitraz and its metabolites
Beta-adrenergic octopamine receptor (BAOR)
major binding site of octopamine, an essential neurotransmitter in arthropods
Amitraz metabolites also bind and result in loss of function or death
Amitraz-resistance SNPs found in
2010 sequenced the gene in resistant and susceptible ticks in Australia and found common mutation in the BOAR of resistant tick populations
Amitraz-resistance SNPs
2 nearby that confer resistance and 2 in a different transmembrane domain
Haplotypes definition
any combination of alleles at multiple loci on the same chromosome
Having the same halotype means
Two homologous chromosomes share the same allele at each of the loci in question
Having the same halotype means
two homologous chromosomes differ at any one of the loci in question
Distribution of haplotypes on a geographical basis is
very information about evolution history
Haplotypes are created by
Combined SNP, microsatellites and indels at nearby loci (or even anywhere within a single chromosome)
Cryptosporidium parvum haplotypes
pathogenicity is closely linked with haplotypes at several loci.
Multi-locus sequence type
Gene-pool defintion
the total number of all alleles (usually considered in relation to a specific gene or genes)
At each locus
there can be one of several different alleles
Microsatellites for most species and loci
contain somewhere between 5-20 different allelic variants
SNPs contain
2 possible allelic variants
Based on gene frequencies, in an equilibrium population, assuming an infinite population size, no migration, no mutation, no selection and no assortative mating (lots of assumptions that are usually wrong) it is possible to predict
genotype frequencies using the Hardy-Weinberg Law.
Does little genetic diversity always come from inbreeding
Nope
The unit square represents
the total population of gametes
ns meaning
the slight difference from expectation is not likely to be statistically significant
Heterozygosity of the next generation depends on
Heterozygosity of the current generation and the size of the population
Rarer alleles in a population likely will trend to
Be removed from future populations
Hardy Weinberg Law
D + H + R = 1
HWE tests
do not detect mutation
don’t detect the effects of low levels of drift, selection, and migration
Indices for genetic diversity: P
Percentage of polymorphic loci
Indices for genetic diversity: A
Alleles per locus
Indices for genetic diversity: H
heterozygosity
Indices for genetic diversity: NE
Effective population size
Observed Heterozygosity (Ho):
simply the proportion of all individuals in a population that is heterozygous at the locus in question
Heterozygosity: N
Number of animals in population
Heterozygosity: NH
Number of heterozygotes
Heterozygosity
Ho = NH/N
Expected Heterozygosity (He) definition
would be expected that if the frequencies of the alleles are similar to each other, then heterozygosity would be higher than if one allele is much more frequent than the other
Expected Heterozygosity
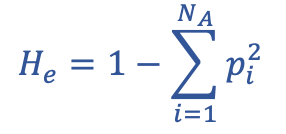
Expected Heterozygosity: NA
number of different alleles
Expected Heterozygosity: p
frequency of the homozygote for each allele
Alleles per locus
impossible to determine at a give locus in a population without testing entire population
Number of Alleles for SNPs
usually 2 but theoretically could be 4
Number of Alleles for microsatellites
varies, but better markers have 10-15 alleles
Allelic richness
interchangeably with the mean number of alleles at any locus OR
enables compensation for N, the number of individuals tested from the population
Measuring overall genetic diversity with alleles
allele that is rare contributes much less to diversity than an allele that has a high frequency
Heterozygosity is high a any locus in a population when
allele frequencies are equal
many alleles are present at the locus in question
Lower than expected levels of heterozygosity strongly suggest
the random sampling effects of genetic drift (including founder effect) have been strong or there is assortative mating
Alleles at any locus over sufficient time is expected to
either be lost from or become fixed at that locus in that population
Wright’s equation
predicts the change in the frequency of heterozygotes (H) from one generation (g) to the next (g+1) for a population of N individuals:
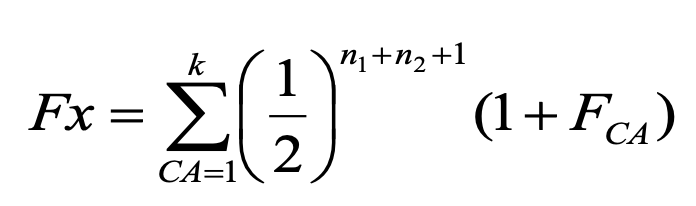
Heterozygosity is always expected to
decline in a closed population without selection
Real populations of animals
don’t usually conform to the assumptions of random selection of gametes and random mating
Animals tend to choose mates
that are more similar to them (positive) or more different (negative)
Sex ratios often deviate from 0.5 and mating is assortative (+ or -)
Effective population size definition
the size of the ideal population that would lose heterozygosity at the same rate as an actual population of interest
Effective population size
very sensitive to deviations from a 0.5 sec ratio and is estimated with equation