MCB4304 MODULE 2 Chromosome Structure and Cloning
1/268
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
269 Terms
What are chromosomes composed of?
Chromosomes are complexes of DNA and proteins.
What does the genome refer to in prokaryotes?
In prokaryotes, the genome typically refers to a single circular chromosome.
How is the genome defined in eukaryotes?
In eukaryotes, the genome refers to one complete set of nuclear chromosomes.
What additional genomes do eukaryotes possess?
Eukaryotes possess a mitochondrial genome and plants also have a chloroplast genome.
What is the main function of genetic material?
The main function is to store the information required to produce the traits of an organism.
What are protein-coding genes responsible for?
Protein-coding genes are responsible for synthesizing RNA and cellular proteins.
What are the necessary DNA sequences for?
They are necessary for the synthesis of RNA, replication of chromosomes, proper segregation of chromosomes, and compaction of chromosomes.
What is the typical structure of prokaryotic chromosomal DNA?
Prokaryotic chromosomal DNA is usually a circular molecule that is a few million nucleotides in length.
How many base pairs does Escherichia coli have?
Escherichia coli has approximately 4.6 million base pairs.
How many base pairs does Haemophilus influenzae have?
Haemophilus influenzae has approximately 1.8 million base pairs.
What accounts for the majority of bacterial DNA?
Protein-coding genes account for the majority of bacterial DNA.
What are intergenic regions?
Intergenic regions are the non-transcribed DNA between adjacent genes.
What is a key feature of prokaryotic chromosomes regarding their structure?
Most prokaryotic species contain circular chromosomal DNA.
How many types of chromosomes do most prokaryotic species contain?
Most prokaryotic species contain a single type of chromosome, which may be present in multiple copies.
What is required to initiate DNA replication in prokaryotes?
At least one origin of replication is required to initiate DNA replication.
What is the nucleoid?
The nucleoid is the region of the cell where the prokaryotic chromosome is found, and it is not bounded by a membrane.
How is the chromosomal DNA compacted in bacteria?
The chromosomal DNA must be compacted about 1000-fold, involving the formation of loop domains.
What are loop domains in bacterial chromosomes?
Loop domains (microdomains) are typically 10,000 bp and help compact the DNA.
How many microdomains are expected in E. coli?
E. coli is expected to have 400 to 500 microdomains.
What are macrodomains in E. coli?
Adjacent microdomains in E. coli are further organized into macrodomains of 800 to 1000 kbp.
What is the relationship between core and microdomains in bacterial chromosomes?
Core and microdomains are structural features that help organize the bacterial chromosome.
What are nucleoid-associated proteins (NAPs) and their functions in bacteria?
NAPs are DNA-binding proteins that help form microdomains and macrodomains, facilitate chromosome compaction and organization, bend DNA or act as bridges for DNA binding, assist in chromosome segregation, and play a role in gene regulation.
How do the structures of archaeal chromosomes vary?
The structure of archaeal chromosomes depends on the DNA-binding proteins they express, with some producing bacterial-like NAPs and others producing eukaryotic-like histone proteins.
What is the role of histone proteins in archaeal chromosomes?
Histone proteins wrap DNA to form nucleosomes and organize it into loop domains, with the number of histones varying among different archaeal species.
What is DNA supercoiling and how does it affect bacterial chromosomes?
DNA supercoiling is the additional coiling of DNA strands due to twisting forces, which further compacts bacterial chromosomes.
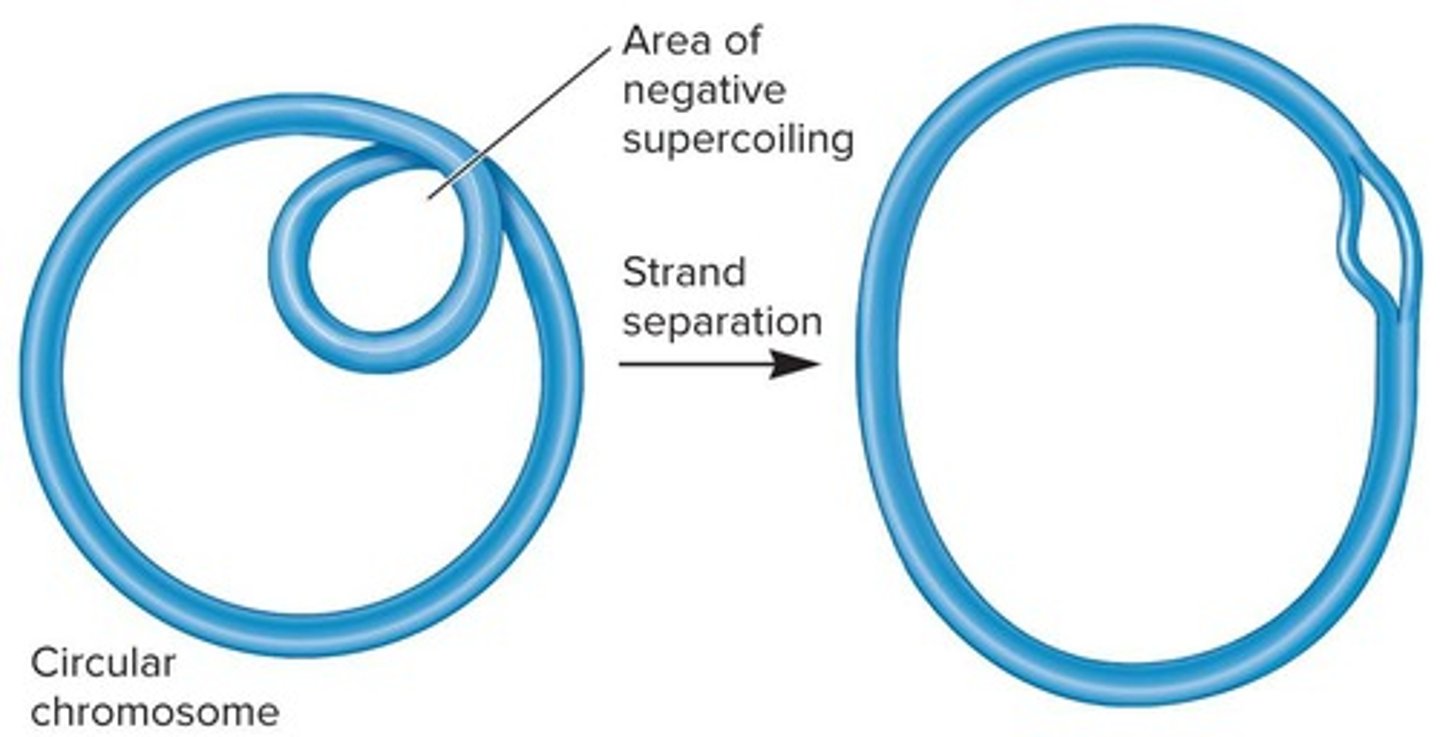
What are the two types of DNA supercoiling?
Underwinding, which can cause fewer turns or negative supercoiling, and overwinding, which can result in more turns or positive supercoiling.
What is the significance of negative supercoiling in bacterial DNA?
Negative supercoiling helps compact the chromosome and creates tension in localized regions that may facilitate DNA strand separation.
What is the relationship between E. coli and negative supercoiling?
In E. coli, there is one negative supercoil per 40 turns of the DNA double helix.
What are the two main enzymes that control DNA supercoiling in bacteria?
DNA gyrase (topoisomerase II), which introduces negative supercoils, and DNA topoisomerase I, which relaxes negative supercoils.
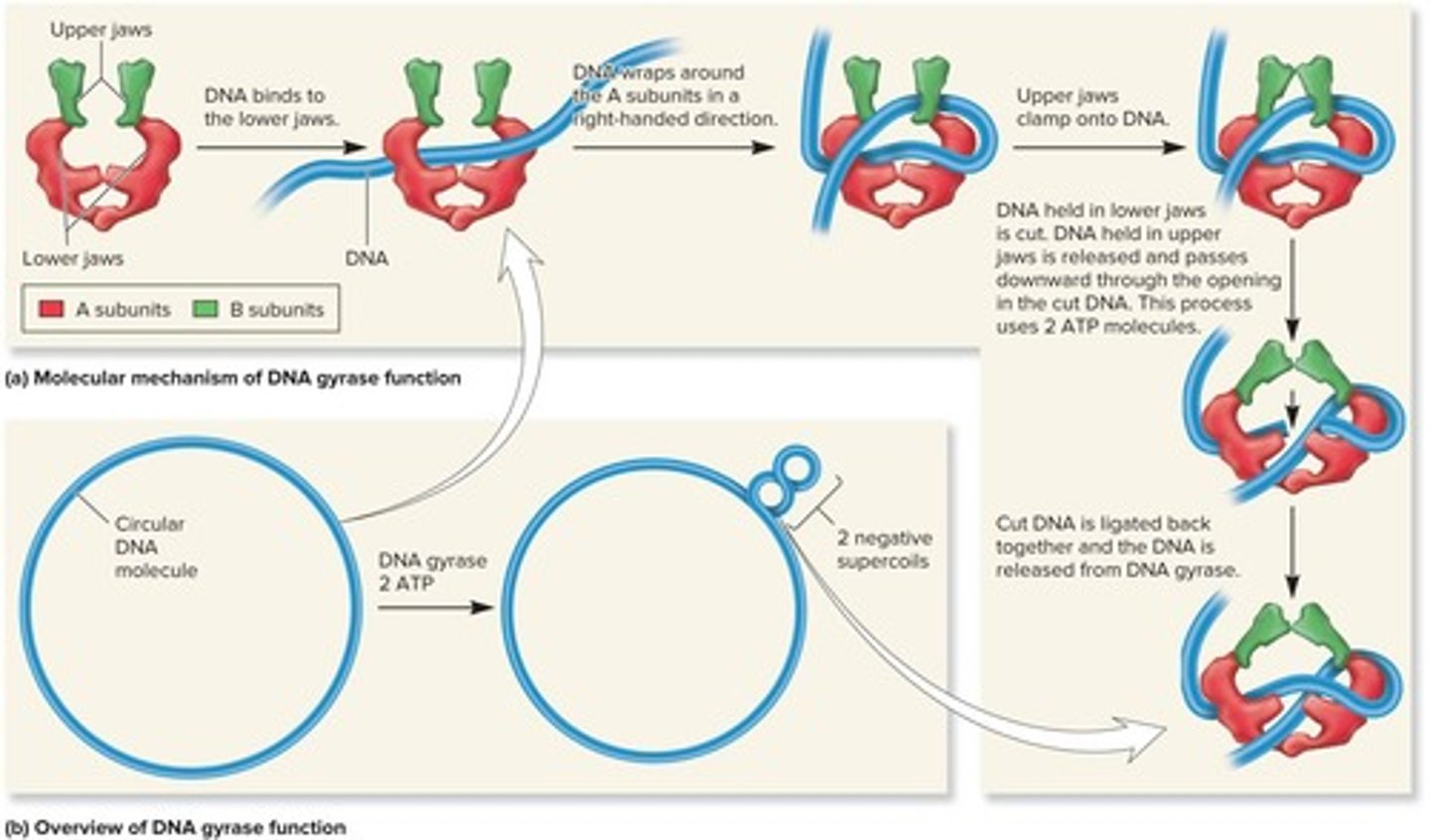
How does DNA gyrase function?
DNA gyrase introduces negative supercoils using energy from ATP, can relax positive supercoils, and untangles intertwined DNA molecules.
What is the effect of inhibiting DNA gyrase in bacteria?
Inhibiting DNA gyrase is a method to cure or alleviate bacterial diseases.
What are the two main classes of drugs that inhibit DNA gyrase?
Quinolones and other bacterial topoisomerase inhibitors.
What is the effect of DNA supercoiling on chromosome function?
DNA supercoiling influences the compaction of chromosomes and promotes strand separation in localized regions.
What happens when DNA is given a turn that unwinds the helix?
It can cause fewer turns or result in the formation of a negative supercoil.
What happens when DNA is given a turn that overwinds the helix?
It can lead to either more turns or the formation of a positive supercoil.
What are topoisomers in the context of DNA supercoiling?
Topoisomers are different forms of DNA that result from supercoiling, including negative and positive supercoils.
How do archaeal species differ in their chromosomal structure?
Some archaeal species produce bacterial-like nucleoid-associated proteins, while others produce eukaryotic-like histone proteins.
What is the impact of negative supercoiling on DNA strand separation?
Negative supercoiling creates tension that promotes strand separation.
How does the action of DNA topoisomerase I differ from that of DNA gyrase?
DNA topoisomerase I relaxes negative supercoils, while DNA gyrase introduces negative supercoils.
Why is the ability of gyrase to introduce negative supercoils crucial for bacteria?
It is essential for bacterial survival, as it facilitates proper DNA compaction and function.
What is the role of DNA-binding proteins in the organization of archaeal chromosomes?
They determine the structure of the chromosome, influencing how DNA is compacted and organized.
What is the significance of loop domains in archaeal chromosomes?
Loop domains help organize DNA within the nucleus, similar to the structure seen in eukaryotic chromatin.
What is an example of a quinolone and its use?
Ciprofloxacin (Cipro), used in the treatment of anthrax and other diseases.
How many sets of chromosomes do humans have?
Humans have 2 sets of 23 chromosomes.
What is the typical structure of a eukaryotic chromosome?
Each chromosome contains a single, linear molecule of DNA, typically with tens to hundreds of millions of base pairs.
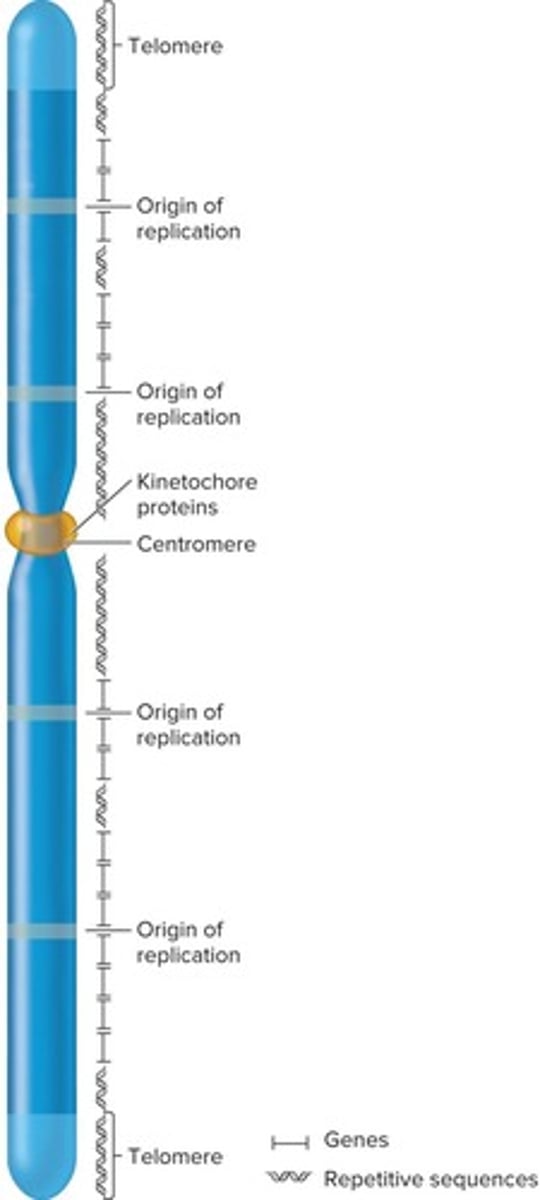
What is the typical number of genes in a eukaryotic chromosome?
A typical chromosome contains a few hundred to several thousand different genes.
How do gene lengths differ between simpler and more complex eukaryotes?
In simpler eukaryotes (like yeast), genes are relatively short; in complex eukaryotes (like mammals), genes are longer and have many introns.
What are the three types of DNA sequences required for chromosomal replication and segregation?
Origins of replication, centromeres, and telomeres.
What is the role of centromeres in eukaryotic chromosomes?
Centromeres play a role in the segregation of chromosomes during cell division.
What are telomeres and their importance?
Telomeres are specialized regions at the ends of chromosomes important for replication and stability.
What is the typical length of eukaryotic chromosomes?
Eukaryotic chromosomes are typically tens of millions to hundreds of millions of base pairs in length.
What is the significance of origins of replication in eukaryotic chromosomes?
Origins of replication are necessary to initiate DNA replication and are found interspersed about every 100,000 base pairs.
What is a characteristic of eukaryotic chromosomes regarding their structure?
Eukaryotic chromosomes are usually linear and occur in sets.
What is the relationship between genome size and species complexity in eukaryotes?
Eukaryotic genomes vary substantially in size, and this variation is not always related to the complexity of the species.
What can cause differences in genome size among closely related species?
Differences in genome size can be due to the accumulation of repetitive DNA sequences rather than extra genes.
What is the genome size of the plant Tmesipteris oblanceolata?
The plant contains 160 billion base pairs.
What is the role of repetitive sequences in eukaryotic chromosomes?
Repetitive sequences are commonly found near centromeric and telomeric regions, and may also be interspersed throughout the chromosome.
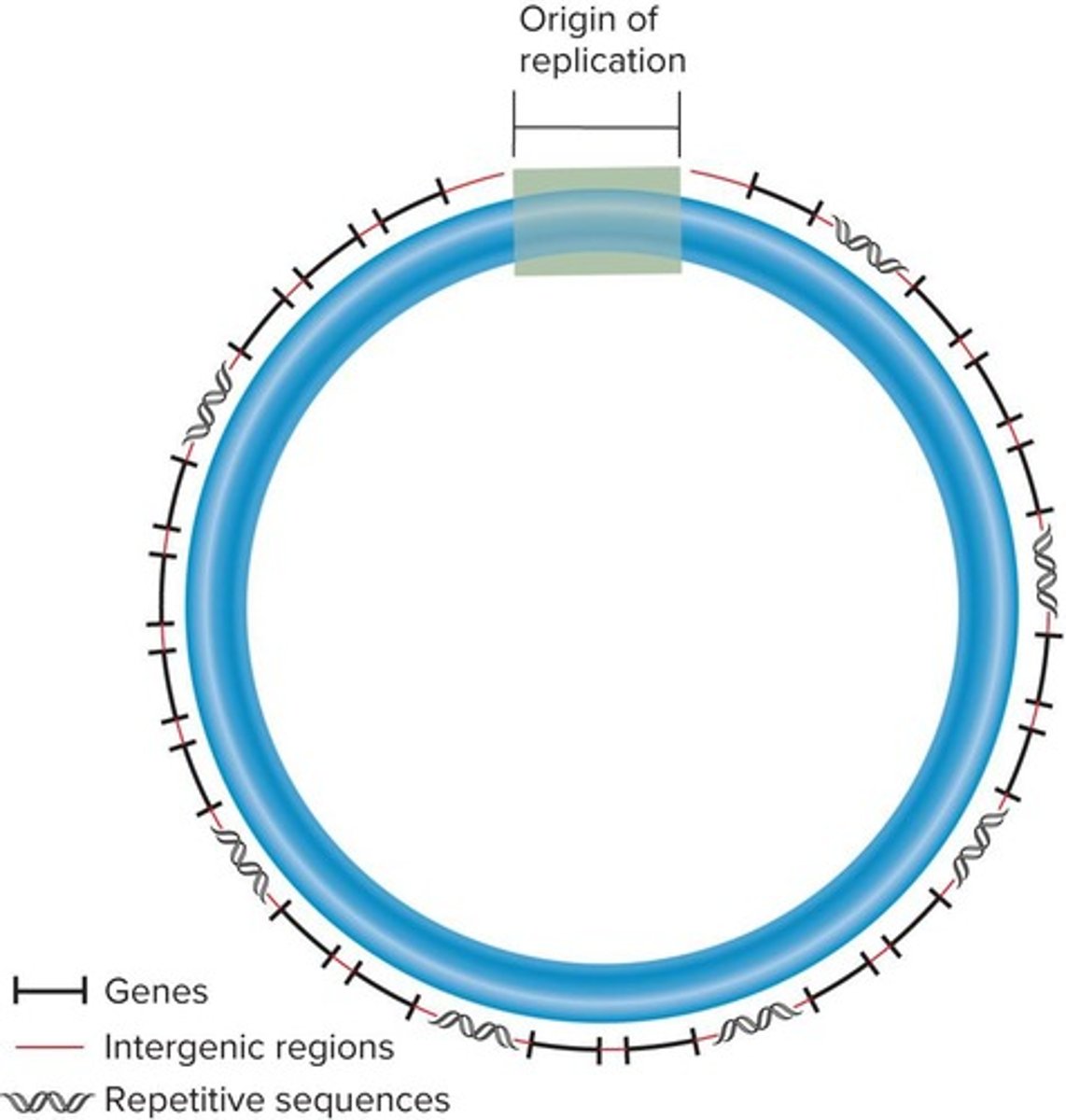
How do eukaryotic chromosomes facilitate the linkage to the spindle apparatus during cell division?
Each chromosome contains a centromere that forms a recognition site for kinetochore proteins.
What is the typical gene length in simpler eukaryotes like yeast?
Genes in simpler eukaryotes are typically several hundred base pairs long.
What is the significance of the number of origins of replication in eukaryotic chromosomes?
Eukaryotic chromosomes contain many origins of replication to ensure efficient DNA replication.
What is the difference in gene structure between complex and simpler eukaryotes?
Complex eukaryotes have longer genes with many introns, while simpler eukaryotes have shorter genes.
What is the typical range for intron lengths in complex eukaryotes?
Intron lengths can range from less than 100 to more than 10,000 base pairs.
What is the general relationship between eukaryotic genome size and the number of genes?
Eukaryotic species typically contain many more genes than bacterial cells.
What does sequence complexity refer to in the context of genomes?
The number of times a particular base sequence appears in the genome.
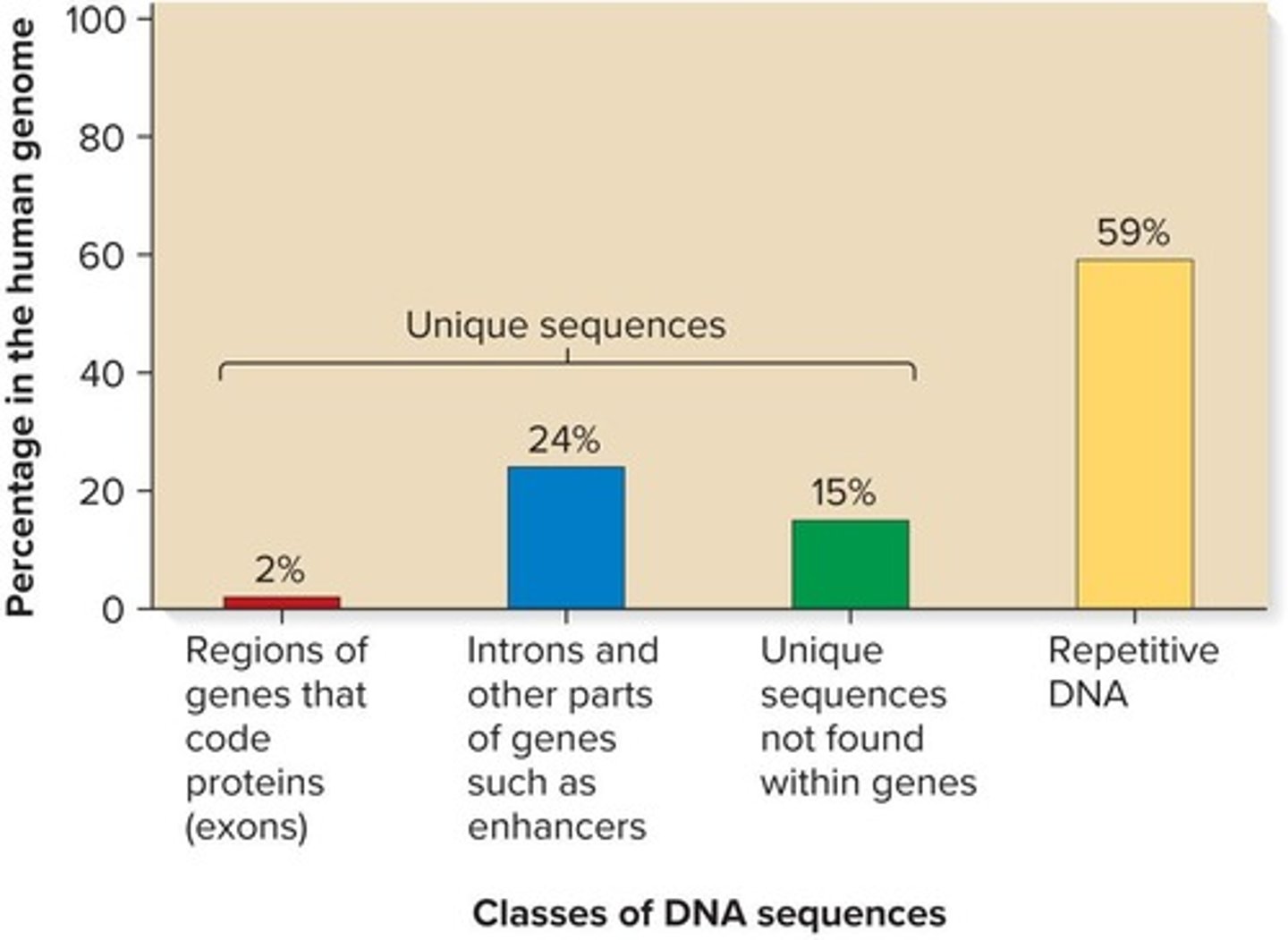
What are the three main types of repetitive sequences in genomes?
Unique or non-repetitive, moderately repetitive, and highly repetitive.
What characterizes unique or non-repetitive sequences?
They are found once or a few times in the genome and include protein-encoding genes and intergenic regions, making up roughly 41% of the human genome.
How many times are moderately repetitive sequences found in the genome?
They are found a few hundred to several thousand times.
What types of sequences are included in moderately repetitive sequences?
Genes for rRNA, histones, sequences that regulate gene expression and translation, and transposable elements.
What defines highly repetitive sequences?
They are found tens of thousands to millions of times, are relatively short, and some are interspersed throughout the genome.
What is an example of a highly repetitive sequence in humans?
The Alu family, which is approximately 300 bp long and represents 10% of the human genome.
How frequently are Alu sequences found in the human genome?
They are found every 5000-6000 bp.
What are tandem arrays of sequences, and where are they commonly found?
Sequences like AATAT and AATATAT are clustered together in tandem arrays, commonly found in centromeric regions.
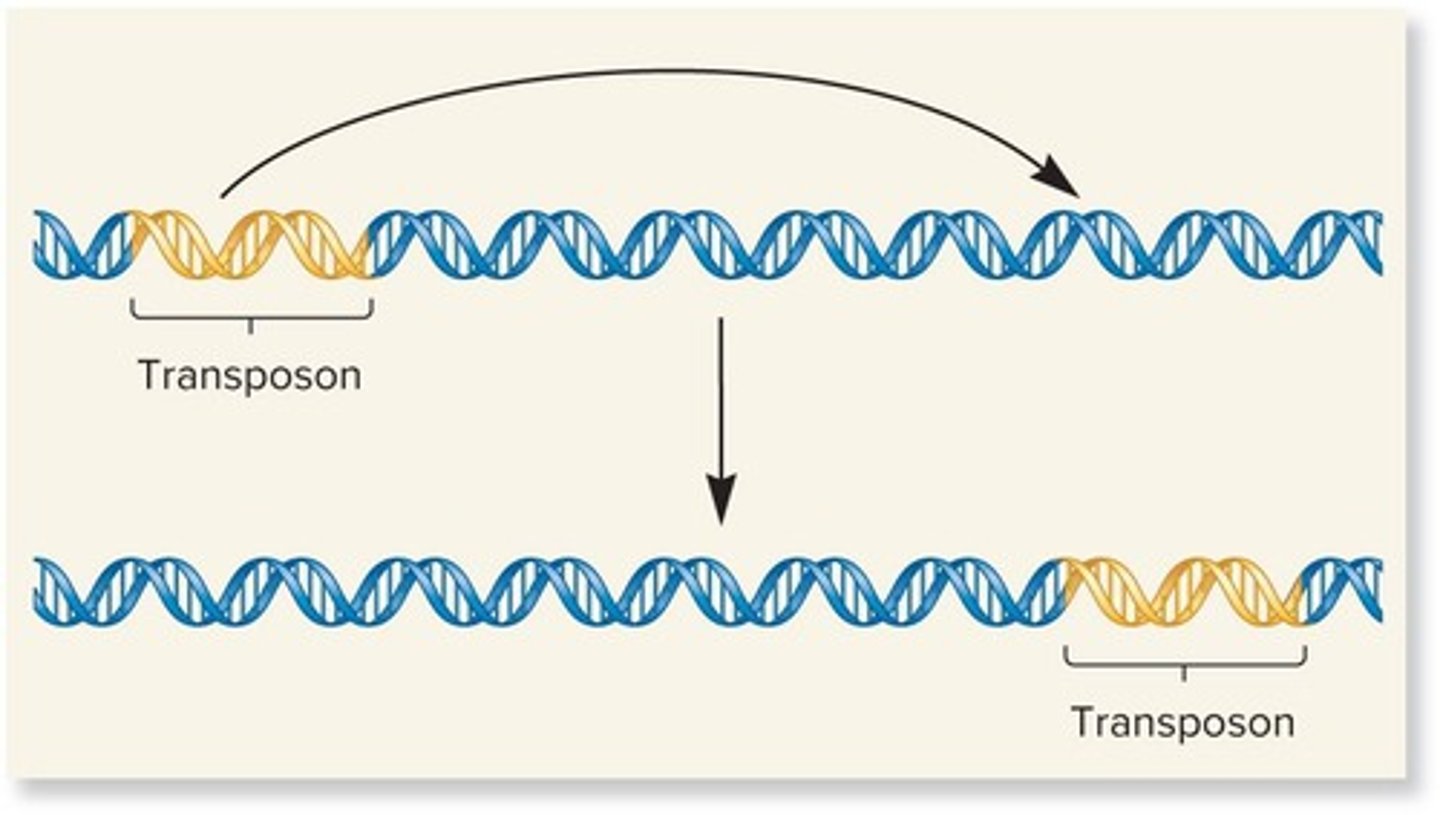
What is transposition in genetic terms?
The integration of small segments of DNA into a new location in the genome.
What are transposable elements (TEs)?
Small, mobile DNA segments that can move within the genome, sometimes referred to as 'jumping genes'.
Who first identified transposable elements, and in what organism?
Barbara McClintock identified TEs in corn in the early 1950s.
What prestigious award did Barbara McClintock receive, and for what discovery?
She received the Nobel Prize in Physiology or Medicine in 1983 for her discovery of mobile genetic elements.
What are the two general types of transposition pathways identified for transposable elements?
Simple transposition and retrotransposition.
What is simple transposition?
A pathway used widely by transposable elements or transposons in bacterial and eukaryotic species.
In what types of organisms have transposable elements been found?
Bacteria, archaea, fungi, plants, and animals.
What percentage of the human genome is made up of highly repetitive sequences?
Approximately 10%.
What is the length range of highly repetitive sequences?
They range from a few nucleotides to several hundred in length.
What role do transposable elements play in the genome?
They can integrate into new locations, potentially affecting gene expression and genome structure.
What is the significance of Barbara McClintock's research on transposable elements?
Her research provided foundational knowledge about genetic mobility and its implications in genetics.
What is the mechanism by which transposable elements (TEs) are transferred to a new target site?
TEs are removed from their original site and transferred to a new target site through a cut-and-paste mechanism.
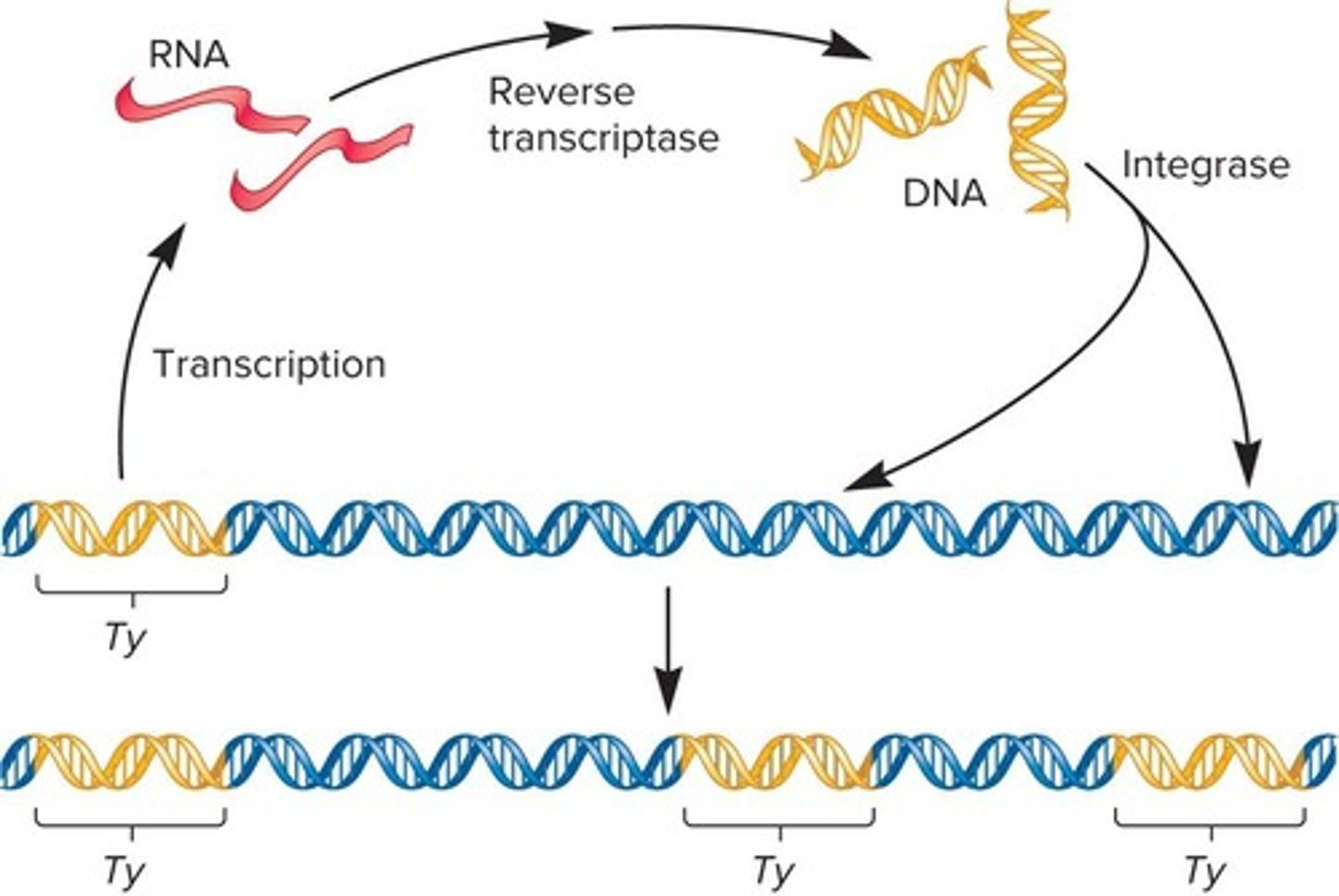
What are retrotransposons and how do they move?
Retrotransposons are transposable elements that move via an RNA intermediate and are transcribed into RNA.
In which types of species are retrotransposons found?
Retrotransposons are found only in eukaryotic species.
What are the two main types of transposition for transposable elements?
The two main types are simple transposition and retrotransposition.
What are direct repeats (DRs) in the context of transposable elements?
Direct repeats are identical base sequences that flank all TEs and are oriented in the same direction.
What is the simplest form of a transposable element?
The simplest TE is an insertion element, which is flanked by inverted repeats.
What are inverted repeats?
Inverted repeats are DNA sequences that are identical or very similar but run in opposite directions.
What is the role of the enzyme transposase in transposition?
Transposase catalyzes the removal of a TE and its reinsertion at another location.
What are LTR retrotransposons and their relationship to retroviruses?
LTR retrotransposons are evolutionarily related to retroviruses and contain long terminal repeats (LTRs) at both ends.
What key proteins do LTR retrotransposons code for?
LTR retrotransposons code for proteins such as reverse transcriptase and integrase, which are needed for retrotransposition.
How do non-LTR retrotransposons differ from LTR retrotransposons?
Non-LTR retrotransposons do not have LTR sequences and may encode proteins that function as both reverse transcriptase and endonuclease.
What is the Alu family of repetitive sequences in humans derived from?
The Alu family is derived from a single ancestral gene known as the 7SL RNA gene.
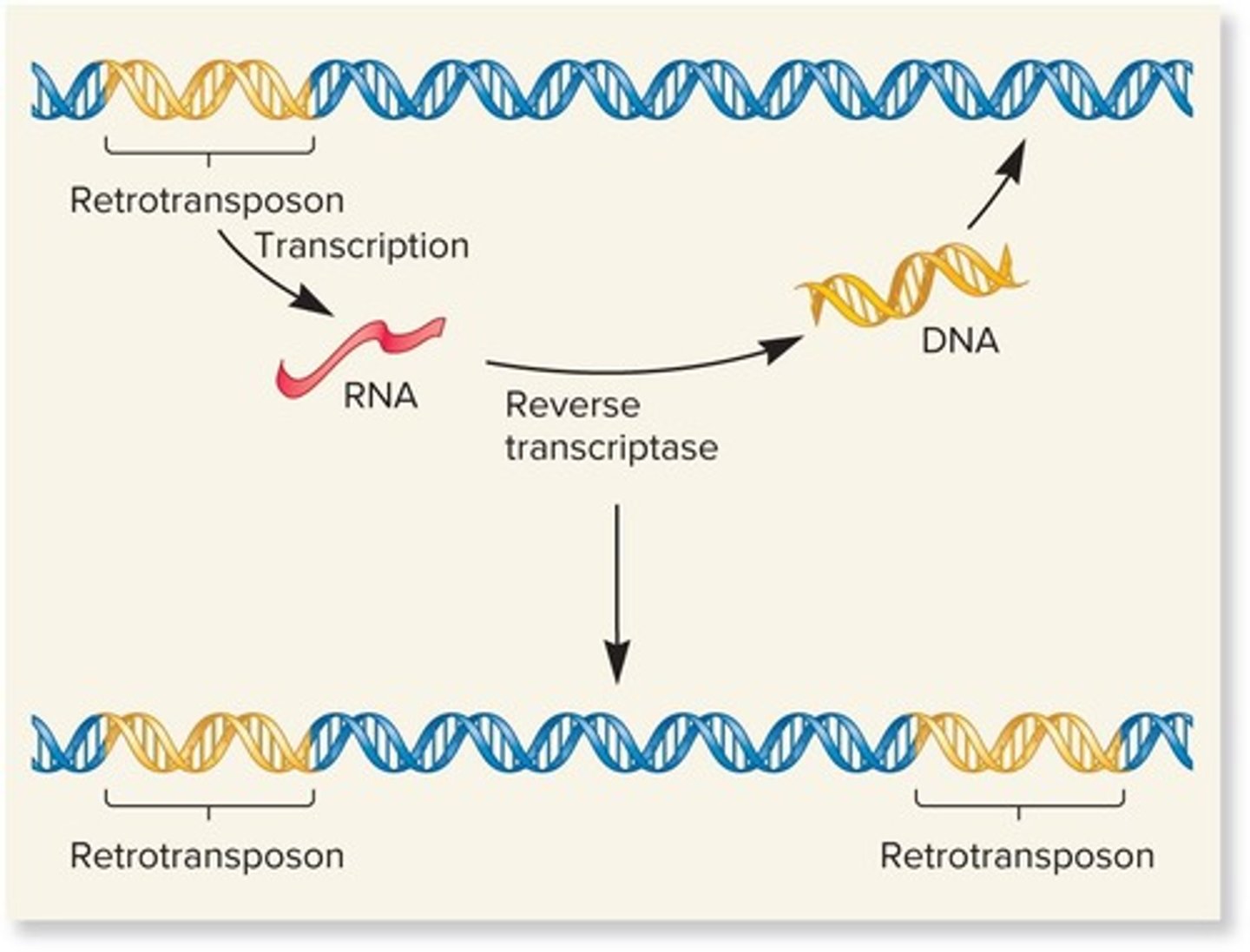
What is the significance of transposition occurring after the replication fork has passed through a TE?
This can lead to an increase in the copy number of the TE, resulting in one chromosome having one TE and another having two copies.
What enzymes are required for LTR retrotransposon movement?
LTR retrotransposon movement requires reverse transcriptase and integrase.
What is the process of target-site primed reverse transcription in non-LTR retrotransposons?
In this process, the retrotransposon is transcribed into RNA, which binds to a nicked site in the target DNA, allowing reverse transcriptase to make a DNA copy.
What is the percentage of the total genome composed of transposable elements in humans?
Approximately 45% of the human genome is composed of transposable elements.
What are LINEs and SINEs in the context of eukaryotic genomes?
LINEs (Long interspersed elements) are usually 1,000 to 10,000 bp long, while SINEs (Short interspersed elements) are less than 500 bp in length.