MT 1 + 2 memory
1/110
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
111 Terms
list the 7 post translational modifications of proteins for bioc 202
phosphorylation
glycosylation
hydroxylation
carboxylation
acetylation
methylation
cleavage
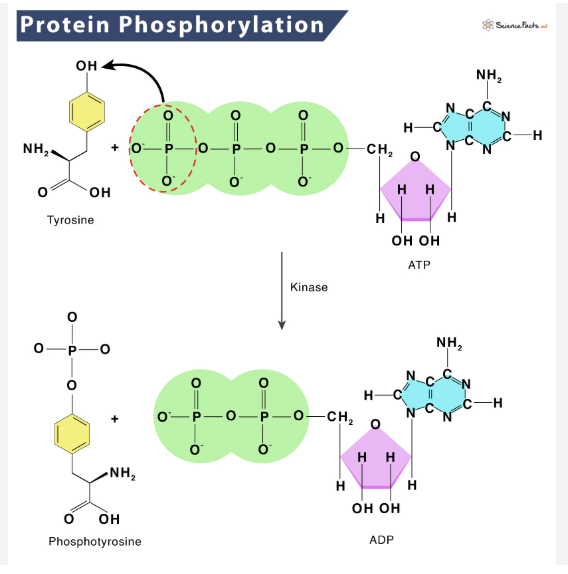
what is phosphorylation and which amino acid residue(s) is it commonly associated with?
phosphate group attaches to -OH
activates or inactivates a protein
associated with: (aa with -OH part of its R group)
Serine, Threonine, Tyrosine
What is glycosylation and which amino acid residue(s) is it commonly associated with?
attachment of one or more sugars to a residue
purposes include surface labelling (most proteins on the cell surface are glycosylated)
associated with: Asparagine, Threonine, Serine and Glutamine (all polar/hydrophillic)
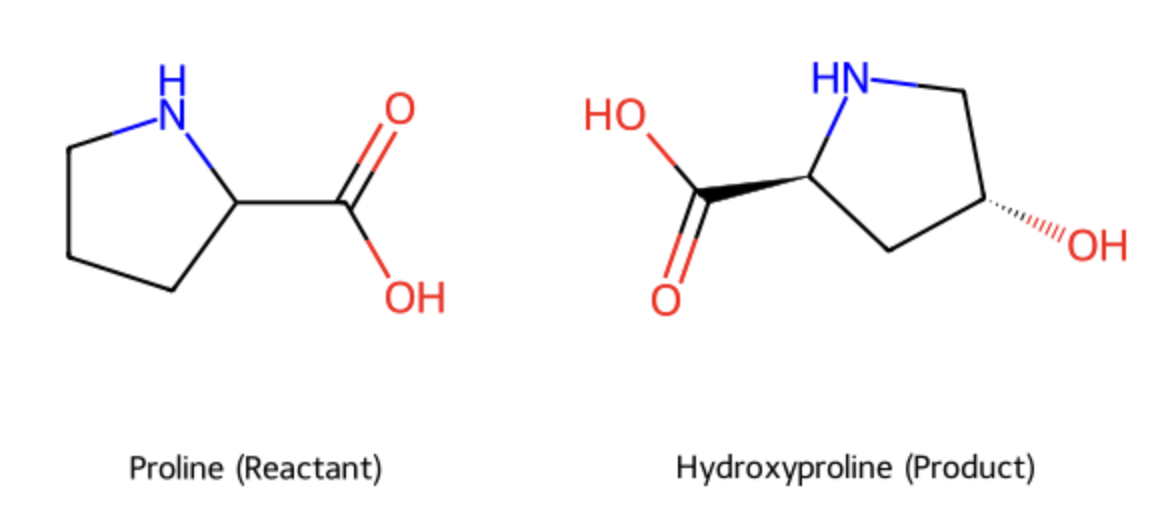
what is hydroxylation and which amino acid residue(s) is it commonly associated with?
addition of an OH (hydroxyl group)
fibre stabilization: vital for collegen
associated with: Proline
what is carboxylation and which amino acid residue(s) is it commonly associated with?
addition of a carboxyl group
usually to Glutamate
clotting
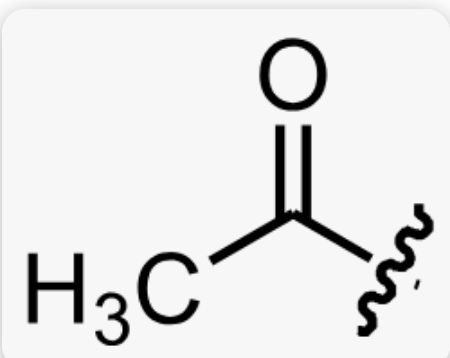
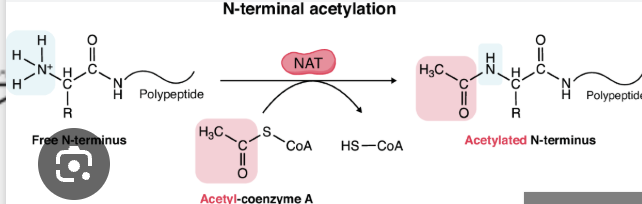
what is acetylation and which amino acid residue(s) is it commonly associated with?
addition of an acetyl group to an NH3+ (amino group)
associated with Lysine
what is methylation and which amino acid residue(s) is it commonly associated with?
the addition of a methyl group to electronegative atoms (N, O)
associated with Arginine, Lysine, Glutamate, Aspartate
what is cleavage and which amino acid residue(s) is it commonly associated with?
most proteins are trimmed after synthesis to activate or inactivate them
multiple proteins from one polypeptide is common
apoenzyme
an enzyme that is inactive because it is missing its cofactor (a required non-protein component)
cofactor
an inorganic ion or a small organic compound required for an enzyme’s activity
holoenzyme
a complete, catalytically active enzyme, consisting of an apoenzyme combined with its necessary cofactor(s)
prosthetic group
a cofactor that is tightly or covalently bound to the active site of an enzyme. A heme group is a common example
transition state
(X‡)
a temporary high energy and unstable state that a substrate must pass through to be converted into a product. It is the point of highest free energy in the chemical reaction (because old chemical bonds are in the process of breaking and new ones are just beginning to form)
also has the lowest concentration
how exactly does an enzyme catalysis work?
the active site of an enzyme acts as a perfect tool for the high-energy unstable “in-between” molecule that exists at the transition state
the enzyme’s active site fits the transition state in two critical ways:
shape: the 3D shape of the active site pocket is precisely arranged to fit the specific bond angles and shape of the transition state molecule
charge: the arrangement of amino acids within the active site creates a specific chemical environment to stabilize it
*by being perfectly suited to hold and stabilize this unstable transition state, the enzyme drastically reduces the amount of energy required to form it, therby reducing the activation energy and speeding up the reaction
activation energy
(ΔG‡)
the minimum amount of energy required for reactants to transform into the transition state
enzymes speed up reactions by lowering this energy barrier
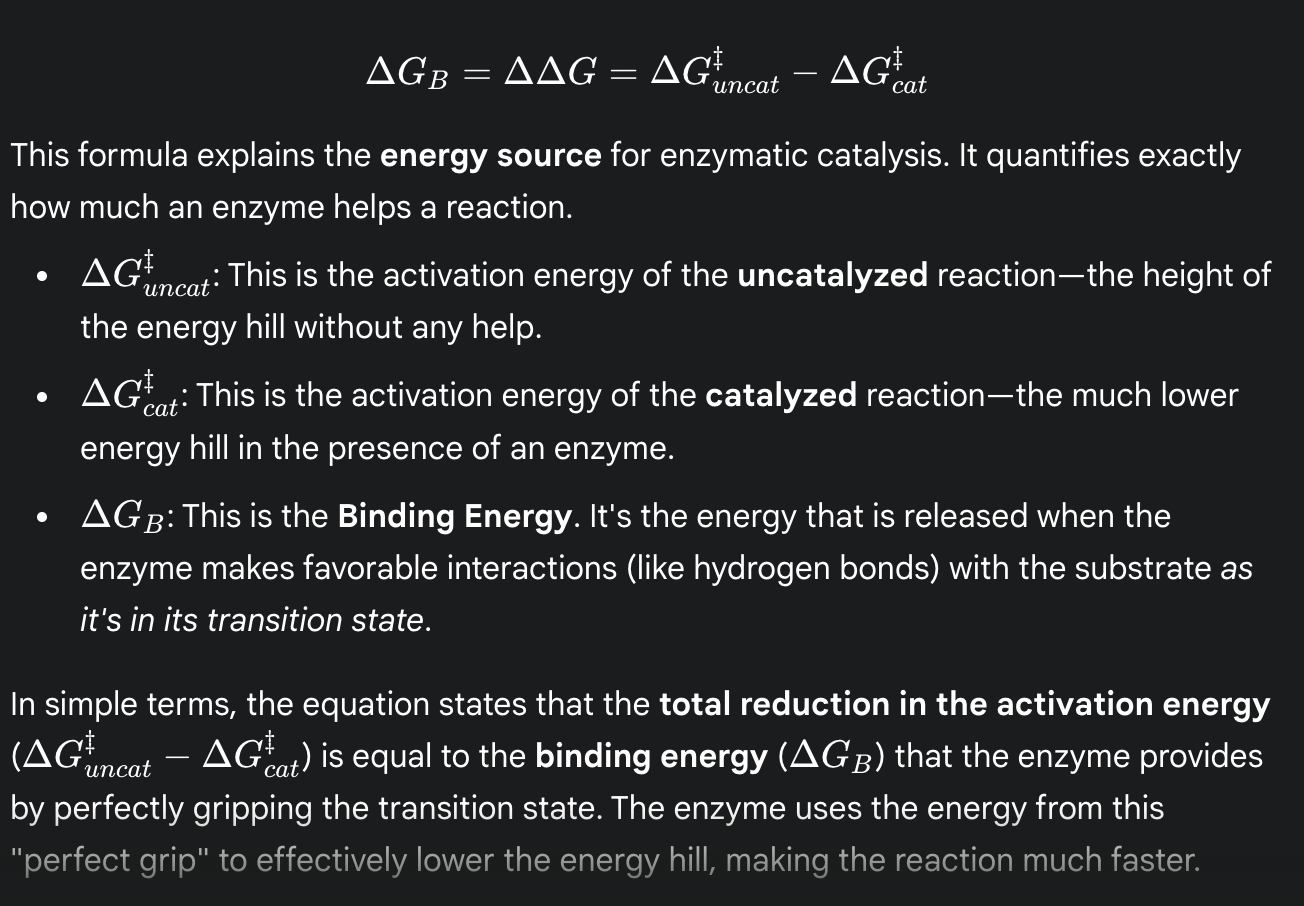
binding energy
(ΔGʙ)
the energy released when an enzyme binds to and stabilizes the transition state
this energy is the primary source for lowering the activation energy
rate of reaction (V, velocity)
The speed at which a reaction proceeds, typically measured as the concentration of product formed per unit of time. Scientists use the initial velocity because at this point, the substrate concentration is at its highest and there is virtually no product yet, which simplifies the measurement and prevents other factors (like the reverse reaction) from influencing the rate.
Rate Constant (k)
a proportionality constant that relates the rate of a reaction to the concentration of the reactants
V0
initial velocity
the reaction rate measured at the beginning of the reaction, when the concentration of the product is negligible
Michaelis-Menten Equation
What It Is: The Michaelis-Menten equation is a mathematical formula that describes the speed (velocity) of a reaction catalyzed by an enzyme. It connects the initial reaction rate (V₀) to the concentration of the substrate ([S]). This allows us to predict how fast an enzyme will work under different conditions.
Where:
V₀ is the initial rate of the reaction.
Vₘₐₓ is the maximum possible rate when the enzyme is completely saturated with substrate.
[S] is the concentration of the substrate.
Kₘ is the Michaelis constant, representing the substrate concentration at which the reaction rate is half of Vₘₐₓ.
Key Shortcut: A crucial concept for solving problems quickly is the definition of Kₘ. If you are told that the reaction is proceeding at exactly half of its maximum speed (V₀ = ½Vₘₐₓ), you immediately know that the substrate concentration must be equal to the Kₘ value ([S] = Kₘ).
![<p><strong>What It Is</strong>: The Michaelis-Menten equation is a mathematical formula that describes the speed (velocity) of a reaction catalyzed by an enzyme. It connects the initial reaction rate (V₀) to the concentration of the substrate ([S]). This allows us to predict how fast an enzyme will work under different conditions.</p><p></p><p>Where:</p><ul><li><p><strong>V₀</strong> is the initial rate of the reaction.</p></li><li><p><strong>Vₘₐₓ</strong> is the maximum possible rate when the enzyme is completely saturated with substrate.</p></li><li><p><strong>[S]</strong> is the concentration of the substrate.</p></li><li><p><strong>Kₘ</strong> is the Michaelis constant, representing the substrate concentration at which the reaction rate is half of Vₘₐₓ.</p></li></ul><p></p><p><strong>Key Shortcut</strong>: A crucial concept for solving problems quickly is the definition of Kₘ. If you are told that the reaction is proceeding at exactly half of its maximum speed (V₀ = ½Vₘₐₓ), you immediately know that the substrate concentration must be equal to the Kₘ value ([S] = Kₘ).</p><p></p>](https://knowt-user-attachments.s3.amazonaws.com/c43256f1-ed65-4b9e-aeb6-831cc1d5ad85.png)
Michaelis Constant (Km)
The substrate concentration required for the enzyme to reach half of its maximum velocity (½ Vₘₐₓ). The Kₘ value is a crucial indicator of an enzyme's affinity for its substrate.
Low Kₘ: Indicates high affinity. The enzyme can work efficiently even at low substrate concentrations.
High Kₘ: Indicates low affinity. The enzyme needs a lot of substrate to be present to work efficiently.
Vmax
The absolute fastest rate at which an enzyme can catalyze a reaction. This speed limit is reached when the enzyme is fully saturated with substrate—meaning every single active site on every enzyme molecule is occupied and working. Adding more substrate at this point will not make the reaction go any faster.
kcat (turnover number)
The maximum number of substrate molecules a single enzyme active site can convert into product per unit of time. It is the most direct measure of an enzyme's catalytic efficiency and represents its intrinsic top speed when fully saturated.
irreversible inhibition
occurs when an inhibitor binds to an enzyme, often covalently and permantely deactivates it.
aspirin for example, irreversibly inhibits the cyclooxyrgenase (COX) enzyme
reversible inhibition
occurs when an inhibitor binds non-covalently to an enzyme, and the enzyme-inhibitor complex can dissociate
this type of inhibition can be overcome
competitive inhibition
a type of reversible inhibition where the inhibitor resembles the substrate and competes for the same active site.
non-competitive inhibition
a type of reversible inhibition where the inhibitor binds to a different site on the enzyme (an allosteric site), changing the enzyme's shape and reducing its activity.
alanine for example acts as a non-competitive inhibitor for the enyme pyruvate kinase
lineweaver-burk plot
a graphical representation of enzyme kinetics, obtained by plotting the reciprocal of the initial velocity (1/V0) against the reciprocal of the substrate concentration. It linearizes the michaelis-menten equation making it easier to determin Vmax and Km
what are the 3 examples of enzymes introduced and what do they do?
papain: speeds up the cleaving of any peptide bond
trypsin: speeds up the cleaving of only the peptide bond on the carboxyl end of Lysine and Arginine
thrombin: speeds up the cleaving of only Arginine-Glycine peptide bonds
characteristics of the active site
they are clefts (aka dimples on the enzyme made up of residues from all over the active site)
occupy a small volume of the enzyme
water is excluded or manipulated from the active site
the substrate is bound to the active site by a series of weak interactions
what is required for an enzyme to function?
it must be partly complementary to its substrate
the general 3D shape of the substrate must match the shape of the active site and the chemical properties (pos, neg, oily, watery region) of the substrate must align with the corresponding properties in the active site
BUT not perfectly…
the substrate and enzyme need a good enough initial fit to bind to each other but the initial binding causes the enzyme to slightly change. its shape, clamping down or wrapping around the substrate more tightly
this creates a more precise and stable connection, perfectly positioning the substrate for the chemical reaction to occur
explain and connect the lock and key and induced binding models
The Lock and Key model is the original, simpler idea. It proposes that the enzyme's active site is a rigid structure that is already a perfect geometric and chemical match for its specific substrate, just like a key fits into its one specific lock.
The Induced Fit model is the modern and more accurate refinement. It states that the enzyme's active site is flexible, not rigid. The initial binding of the substrate is good but not perfect, and this interaction induces the active site to change its shape to wrap more tightly around the substrate.
The connection is that the Induced Fit model builds upon and improves the Lock and Key concept. It keeps the essential idea of a specific match but adds the crucial element of enzyme flexibility. This flexibility is what allows the enzyme to stabilize the reaction's transition state, which is the key to speeding up the reaction.
do enzymes alter the final equilibrium of products to reactants or alter the change of free energy of the reaction?
no
can enzymes change the spontaneity of a reaction, for example make a non-spontaneous reaction with positive change in gibbs free energy spontaneous?
no
is the active site of the enzyme perfectly complementary to the transition state?
yes
what do the following symbols represent? GS, GP, GX‡
GS is the starting energy level of the Substrate.
GP is the final energy level of the Product.
GX‡ is the energy of the Transition State—the highest-energy, most unstable point in the middle of the reaction.
what is the formula for the activation energy for the forward reaction
(the amount of energy needed to get from the starting energy level of the substrate up to the peak of the energy hill)
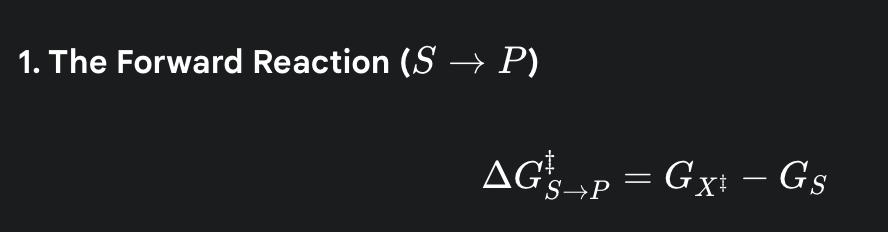
what is the formula for the activation energy for the reverse reaction
(the amount of energy required to get the energy level of the product back up to the same energy hill)
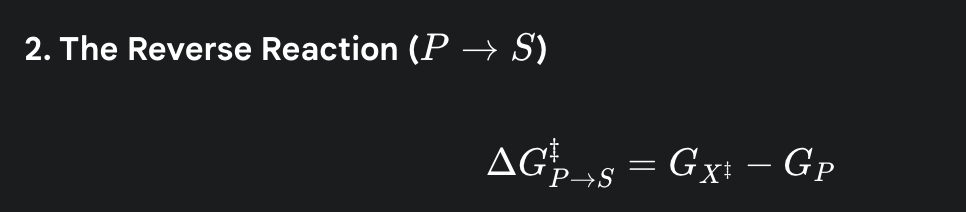
reaction pathway (physical steps of an enzyme reaction)

formula for binding energy
(the total reduction in activation energy is equal to the binding energy that the enzyme provides by perfectly gripping the transition state)
what bond joins amino acids
peptide bonds
what is phi and psi angles?
phi: dihedral angles of N-Ca
psi: dihedral angles Ca-C=O
what are the characteristics of the bonds in a polypeptide chain
C=O to NH bond is locked due to double bond character as a result of resonance; results in same plane
psi and phi bonds (other bonds beside 1) have free rotation
BUT not all combinations of psi and phi angles are possible (combinations defined by ramachandran plot)
special aa + side chains
cysteine: 1c, thiol (disulfide bonding)
selenocysteine: 1c, SeH
proline: 5 mem w/ amino (NP), R config
Glycine: NONE (NP) (achiral)
polar aa + R groups
Asparagine: 1 c, amide
Glutamine: 2 c, amide
Threonine: 2 c, OH branch
Serine: 1 c , OH
hydrophobic (mostly np) aa + R groups
Alanine: 1 c
Valine: 2 c, 1 c branch
Isoleucine: 3 c, 1 c branch (1)
Leucine: 3 c, 1 c branch (2)
Methionine: 2 c, S, 1 c
Phenylalamine: 1 c, phenyl
Tyrosine: 1 c, phenyl, OH (opp starting c)
Tryptophan: 1 c, indole (5 mem NH (3), 6 mem)
which aa have R groups with no carbon chains
glycine and proline
which aa have R groups with 1 carbon chains
cys (2), rings (4), asp (2), alanine + serine
cysteine
selenocysteine
phenylalamine
tyrosine
tryptophan
histidine
asparagine
aspartate
alanine
serine
which aa have R groups with 2 carbon chains
Glu, MVP
Glutamine
Glutamate
Methionine
Valine
Threonine (Polar)
which aa have R groups with 3 carbon chains
Leu (2), Arg
Leucine
Isoleucine
Arginine
which aa has a R group with a 4 carbon chain
Lysine
What are the amino acids for the following 1 letter codes:
R, N, D, Q, E, K, U, Y, W, F, T, A
Arginine, Asparagine, Aspartate, Glutamine, Glutamate, Lysine, Selenocysteine, Tyrosine, Tryptophan, Phenylalamine, Threonine, Alanine
what are the 3 letter codes for the following amino acids :
Glutamate, Aspartate, Glutamine, Asparagine, Isoleucine, Tryptophan, Selenocysteine
Glu, Asp, Gln, Asn, Ile, Trp, Sec
what is the exception to favouring trans peptide bonds
X-Proline
some can be cis bc both have steric hinderance (the energy diff between cis and trans is smaller compared to the diff w other aa)
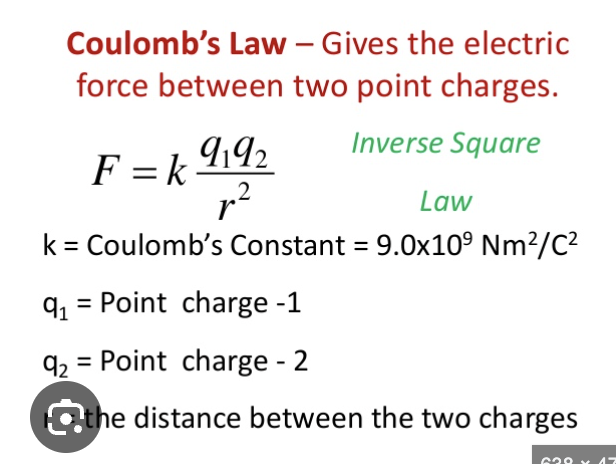
coulomb’s law
pKa
specific pH at which a functional group is exactly 50% protonated and 50% deprotonated (tipping point)
buffering zone is when pKa is within ± 1 of the pH (a certain amount of acid/base needs to be added to overcome this
what are the functional groups among the amino acids
hydrophobic
alkyl (just carbon; alanine, valine, isoleucine, leucine)
phenyl (phenylalamine)
phenol (tyrosine)
indole (tryptophan; 5 mem ring, NH (3) + 6 mem)
thio-ether (methionine: c - s - c)
pyrrolidine (proline)
hydrophilic
hydroxyl (serine, threonine)
amide (asparagine, glutamine)
thiol (cysteine; can form disulfide bonds, weakly H bonds)
SeH
basic (neutral to +)
imidazole (histidine; 5 mem, N (2), NH (3)
guanidinium (arginine; NH, NH, NH2)
side-chain amino (lysine)
acidic (neutral to -)
side chain carboxyl (aspartate, glutamate)
which aa has constrained phi rotation?
proline
strongest interaction among aa
cysteine - cysteine
disulfide bond (covalent)
a-helix wreckers
proline + glycine
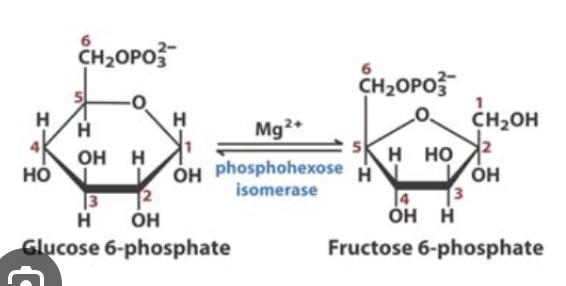
what are the 11 molecules involved in glycolysis?
Glucose
Glucose 6-phosphate
Fructose 6-phosphate
Fructose 1,6-bisphosphate
Dihydroxyacetone phosphate (DHAP)
Glyceraldehyde 3-phosphate (GAP)
1,3 - Bisphosphoglycerate
3 - Phosphoglycerate
2 - Phosphoglycerate
Phosphoenolpyruvate
Pyruvate
(Glu, Glu,
Fru, Fru,
D, G, B (1),
Phos (3), Phos (2), Phos,
Pyruvate)
which reactions are irreversible in the glycolysis pathway?
reaction 1: glucose → glucose 6-phosphate
reaction 3: fructose 1,6-phosphate → fructose 1,6-bisphosphate
reaction 10: phosphoenol pyruvate → pyruvate
(first, third, last)
what are the enzymes involved in the glycolysis pathway?
hexokinase
phosphoglucose isomerase
phosphofructokinase -1 (PFK1)
aldolase
triosephosphate isomerase
glyceraldehyde-3-phosphate dehydrogenase
phosphoglycerate kinase
phosphoglycerate mutase
enolase
pyruvate kinase
(Hey, please pick all the green prickly pears, every pear!)
why is the payoff stage of glycolysis happen twice?
there is a split step
in reaction 4, fructose 1,6 bisphosphate splits into dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (G3P)
but only G3P can be used to further the reaction pathway
but triose phosphate isomerase can quickly convert DHAP into another G3P
because you have two molecule of G3P the payoff stage can happen twice
what are the inputs and byproducts of the glycolysis pathway
reaction 1: - ATP + ADP (glucose → glucose 6-phosphate)
reaction 3: - ATP + ADP (fructose 6-phosphate → fructose 1,6-bisphosphate)
reaction 6: - Pi - NAD+, + NADH (glyceraldehyde 3-phosphate)
reaction 7: - ADP + ATP (1,3 bisphosphoglycerate → 3-phosphoglycerate
reaction 9: + H2O (2-phosphoglycerate → phosphoenolpyruvate)
reaction 10: -ADP + ATP (phosphoenol pyruvate → pyruvate)
end: + 2 ATP, 2 Pyruvate, 2 NADH, 2 H2O
draw glucose and pyruvate
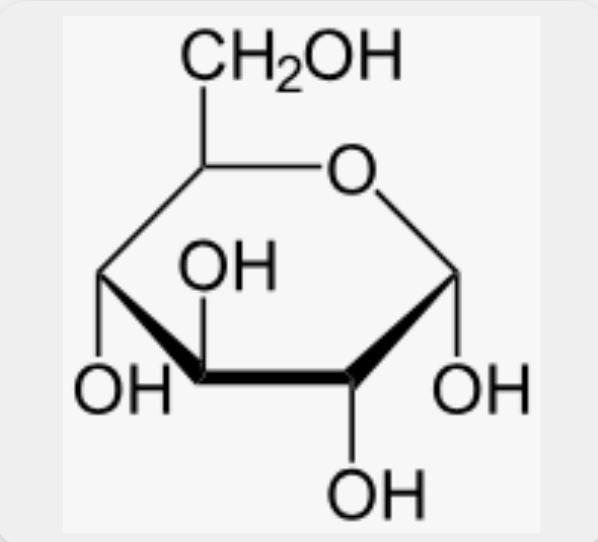
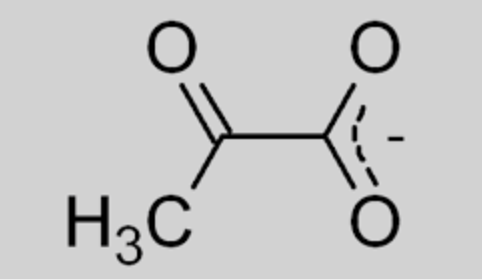
draw: glucose 6-phosphate
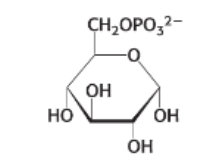
draw: fructose 6-phosphate
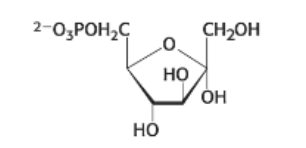
draw: fructose 1,6 - bisphosphate
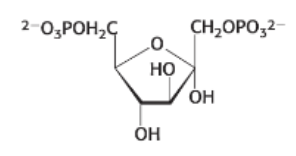
draw: dihydroxyactetone phosphate
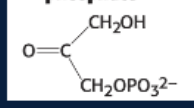
draw: glyceraldehyde 3-phosphate
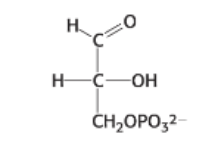
draw: 1,3-bisphosphoglycerate
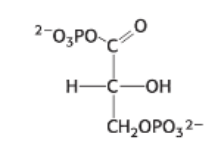
draw: 3-phosphoglycerate
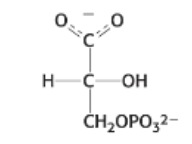
draw: 2-phosphoglycerate
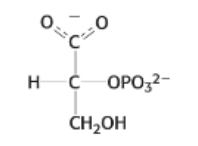
draw: phosphoenolpyruvate
what are the different metabolic pathways that occur after glycolysis and how does the presence or absence of oxygen determine what happens next?
The central theme is the critical need to regenerate NAD+ which is essential input for glycolysis (two NAD+ is consumed to create 2 NADH)
A) Aerobic Conditions (oxygen present)
pyruvate processing: 2 pyruvate molecules are converted into 2 Acetyl-CoA (by the PHD complex). This step also produces more NADH
citric acid cycle: 2 Acetyl-CoA molecules enter the citric acid cycle, generating more NADH, CO2 and H2O
The NAD+ Solution: all the NADH (from glycolysis, PHD and the citric acid cycle) travels to the mitochondrial electron transfer chain. Here NADH unloads its electrons, with oxygen (O2) acting as the final electron acceptor, producing water
result: This process regenerates NADH+ which can then cycle back to keep glycolysis running. This is the most efficient way to make energy (ATP)
B) Anaerobic conditions (no oxygen)
problem: without oxygen the mitochondrial electron transfer chain stops. There is nowhere for NADH to dump its electrons
As a results the cell’s NAD+ gets stuck as NADH. Since glycolysis requires NAD+ as an input, glycolysis stops which means the cell can no longer produce ATP
solution: to solve this, cells uses fermentation
lactate fermentation: (human muscles) pyruvate is converted into 2 Lactate
ethanol fermentation: (yeast) pyruvate is converted into 2 Ethanol and 2 CO2
result: as the red arrows show, both fermentation pathways use the NADH (oxidizing it) to regenerate NAD+ . This NAD+ allows the cell to produce a small but vital amount of ATP even without oxygen
what are these molecules
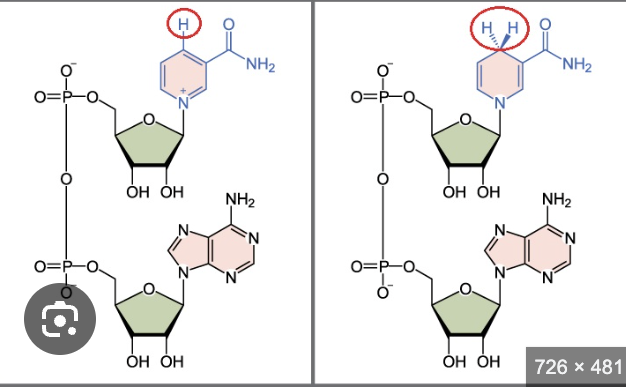
NAD+/NADH
Nicotinamide Adenine Dinucleotide (oxidized form and reduced form)
what are these molecules
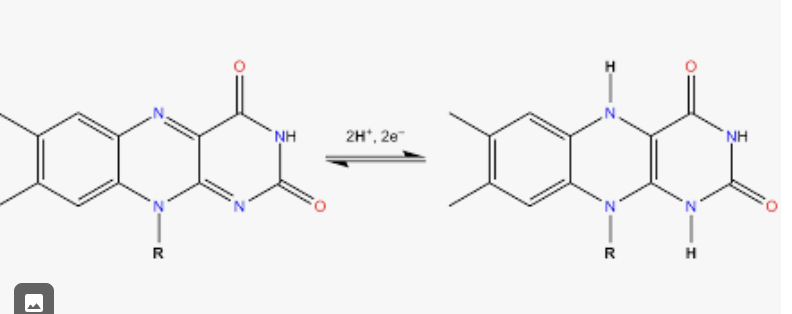
FAD/FADH2
Flavin Adenine Dinucleotide (oxidized and reduced form)
what are these molecules
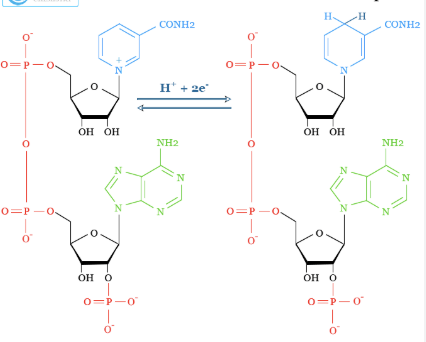
NADP+/NADPH
Nicotinamide Adenine Dinucleotide Phosphate (oxidized and reduced form)
what are these molecules
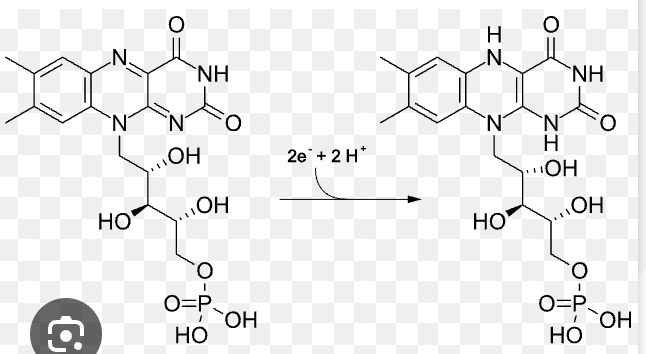
FMN/FMNH2
Flavin Mononucleotide (oxidized and reduced form)
simpler form; part of FAD
what does NAD+ and NADP + stand for? function?
NAD: nicotinamide adenine dinucleotide
NADP: nicotinamide adenine dinucleotide phosphate
can move around the compartment
can move from enzyme to enzyme
able to recognize all different forms
aldose vs ketose. draw example of linear and ring form.
aldose:
a sugar with its carbonyl group at the end of the carbon chain, forming an aldehyde (-CHO)
can be in a ring form when a hydroxyl group (usually on C5) attacks the C1 aldehyde. The resulting ring is called a hemiacetal
typically form 6-membered rings (pyranoses)
ketose:
a sugar with its carbonyl group on an internal carbon (not at the end), forming a ketone *C-CO-C
can be in ring form when a hydroxyl group (usually on C5) attacks the C2 ketone. The resulting ring is called a hemiketal
they typically form 5-membered rings (called furanoses)
The C2 carbon is the anomeric carbon
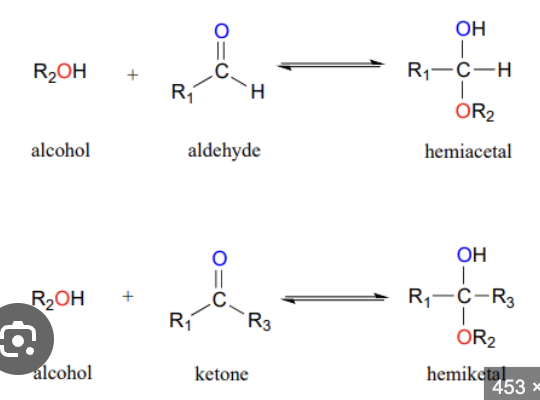

what is a carbonyl group?
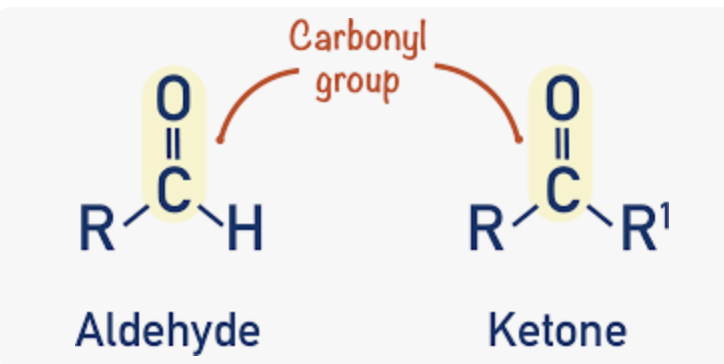
a functional group consisting of one carbon atom double bonded to one oxygen atom; group is highly reactive and is the site of the chemical reaction that causes sugar to form a ring
what is an anomeric carbon?
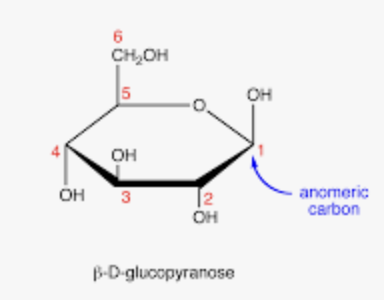
the most important carbon in a sugar ring; the specific carbon that used to be the carbonyl carbon in the sugar’s linear (open-chain) form; a new chiral center
defined as the only carbon atom in the ring to be be bonded to two oxygen atoms (one in the ring and one in the -OH group
for an aldose, the anomeric carbon is C1
for a ketose the anomeric carbon is C2
numbering carbons
determined by the numbering of the original linear (open-chained) form of the sugar
start numbering from the end that gives the carbonyl group (C=O) the lowest possible number
for a 6-membered glucose ring the anomeric carbon is C1
for a 5-membered fructose ring the anomeric carbon is C2
what is a sugar?
chains of carbons that contain mutiple hydroxyl groups and one carbonyl group
monosacharide (glucose or fructose)
disacharides are two sugar units (sucrose or table sugar)
polysacharides are long chains
which reaction is a classic example of induced fit?
reaction 1: glucose → glucose 6-phosphate via hexokinase
enzyme changes shape upon binding glucose
what is the committed step in the glycolysis pathway and what does this mean?
reaction 3: fructose 6-phosphate → fructose 1,6-bisphosphate
point of no return
what is the coupled reaction in the glycolysis pathway?
reaction 6: glyceraldehyde-3-phosphate → 1,3 - bisphosphoglycerate
a highly favourable oxidation of GAP is coupled to the unfavourable addition of a phosphate via a high energy thioester intermediate in the enzyme’s active site
mutase
enzyme that shifts a functional group
which reaction involves dehydration and the creation of water?
reaction 9: 2-phosphoglycerate → phosphoenolpyruvate
what is the net reaction of glycolysis?
Glucose + 2Pi + 2ADP + NAD+ → 2 Pyruvate + 2ATP + 2NADH + 2H+ + 2H2O
how is NAD+ regenerated in anaerobic conditions (no oxygen)?
pyruvate converted to Acetyl-CoA and enters Krebs cycle
NADH travels to electron transport chain where it is oxidized back to NAD+
O2 is the fianl electron acceptor
how is NAD+ is regenerated in anaerobic conditions
lactate fermentation
in mammals
pyruvate reduced to lactate by lactate dehydrogenase
reaction oxidizes NADH back to NAD+, allowing glycolysis to continue
pyruvate itself is the final electron acceptor
ethanol fermentation
in yeast
two-step process
a) pyruvate is converted to acetaldehyde releasing CO2
b) acetaldehyde is reduced to ethanol. This reaction oxidizes NADH back to NAD+
acetaldehyde is the final electron acceptor
where does glycolysis regulation occur? what are the inhibitors and activator of the enzymes involved in regulating?
allosterically at the irreversible steps of the glycolysis pathway
hexokinase
inhibited by: own product, glucose 6-phosphate
regulation does not occur in the liver
phosphofructofinase
inhibted by: high ATP (signals that energy is abundant), high citrate (an intermediate of the krebs cycle; presence signals that the downstream cycle is full)
activated by: high AMP (a much more sensitive indicator of low energy than ADP), fructose 2,6-bisphosphate (a hormonal signal in the liver)
pyruvate kinase
inhibted by: high ATP, Alanine (an amino acid synthesized from pyruvate)
activated by: fructose - 1, 6-bisphosphate (the product of PFK); this is feed-forward stimulation where a product from an earlier step activates an enzyme later in the pathway.
what is the effect of an increased enzyme concentration of Vmax and Km
Vmax increases (directly proportional to the number of workers/enzyme concentration)
Km does not change because it is a measure of the enzyme’s affinity for its substrate (how sticky is the active site) and not a measure of speed
A low Km means that the active site is very sticky and finds its substrate easily (high affinity)
How is Vmax, kcat and Km affected by irreversible, reversible competitive, and reversible non-competitive inhibition
irreversible competition: Vmax decreases, kcat decrease, Km unchanged
reversible competitive inhibition: Km increases (it takes more substrate to outcompete the inhibitor), but Vmax is unchanged
reversible non-competitive inhibition: Vmax decreases but Km is unchanged
pyranose and furanose
pyranose
a sugar that has formed a six membered ring structure consiting of five carbon atoms and one oxygen atom
most common and stable ring form for six carbon aldoses like glucose
furanose
a sugar that has formed a five-membered ring structure, consisting of four carbon atoms and one oxygen atom
common ring form for ketoses like fructose
NAD+ and NADH
two forms of the same crucial molecule, Nicotinamide Adenine Dinucleotide
the cell’s “rechargeable “energy battery” or “shuttle”
main job is to act as a coenzyme that carries electrons from one reaction to another
NAD+: oxidized form; ready to accept high-energy electrons (and protons) from a substrate that is being oxidized
in glycolysis NAD+ accepts electrons and becomes NADH
NADH: reduced form; carries two high-energy electrons and protons (NADH + H+); NADH then transports these electrons to another location. In aerobic respiration, it donates them to the electron transport chain to make a large amount of ATP. In anaerobic fermentation, it donates them to pyruvate to regenerate NAD+, allowing glycolysis to continue
*constant cycling between NAD+ and NADH (redox) is essential for converting the energy in food (like glucose) into a useable form (ATP)
describe the chymotrypsin mechanism
substrate binds the active site and the large hydrophobic residue is recognized by the hydrophobic pocket
Ser 195 does a nucleophillic attack on the carbonyl carbon of the substrate; proton from Ser 195 is transferred to His 57
This causes the carbon to adopt a tetrahedral geometry forming an oxyanion; this oxyanion is stabilized by H-bonding from the enzyme backbone in the oxyanion hole
the tetrahedral intermediate rearanges to the acyl-intermediate, cleaving the peptide bond while His 57 acts as a acid catalyst (donates a proton) to the amine group to promote degradation
The new amino peptide fragment (product 1) leaves
A water molecule enters
water’s partial negative charge acts as a nucleophile attacking the carbonyl carbon and in the process, donating a proton to His
This forms a second oxyanion tetrahedral intermediate
The His acts as an acid catalyst donating the protein back to Ser 195 dorming the new carboxylic acid and free Ser 195
The new carboxylic acid peptide fragment (product 2) leaves and the enzyme is rejuvenated
*note: (His 57 has both accepted and donated a proton ) x2