Bio 1b Evolution Section II: Phylogenetics
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
What are the 3 goals of systematics? Be sure to mention phylogenetics.
Identifying and naming species
Establishing the evolutionary relationships between taxa (phylogenetics)
Phylogenetics: Determining the evolutionary relationships between species (and higher taxa)
Developing classifications
What is a cladogram?
diagrams that depict the relative degree of relatedness among taxa, nothing more
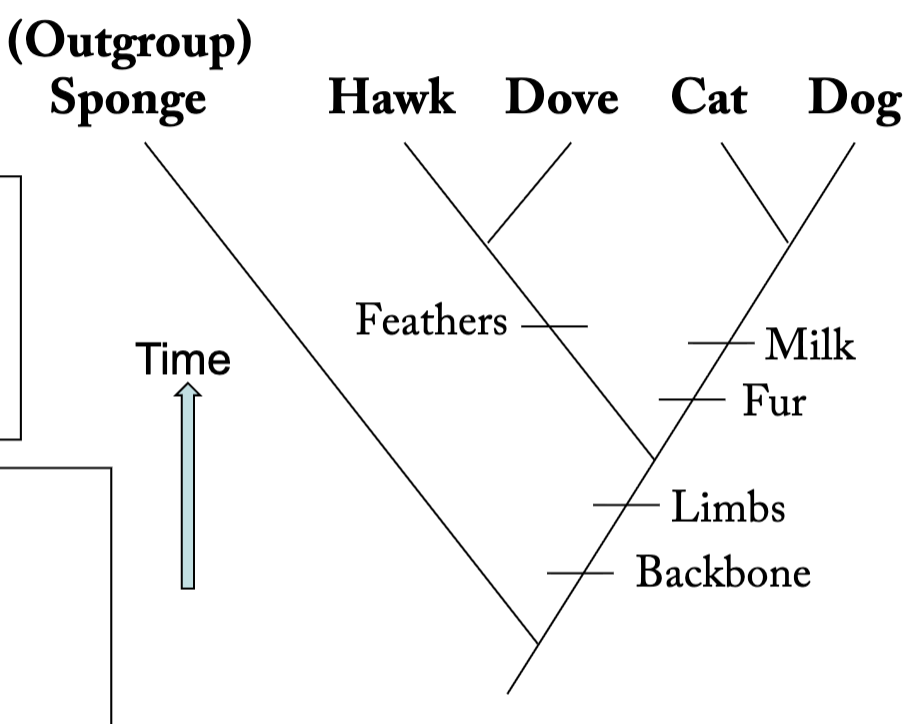
How are cladograms made? Be sure to mention the character matrix and the principle of parsimony.
Taxa are grouped based on shared evolutionary innovations, termed synapomorphies (e.g., fur is a synapomorphy of the Cat and Dog).
The primitive character state is said to be plesiomorphic, if shared it is a symplesiomorphy. The derived state is said to be apomorphic
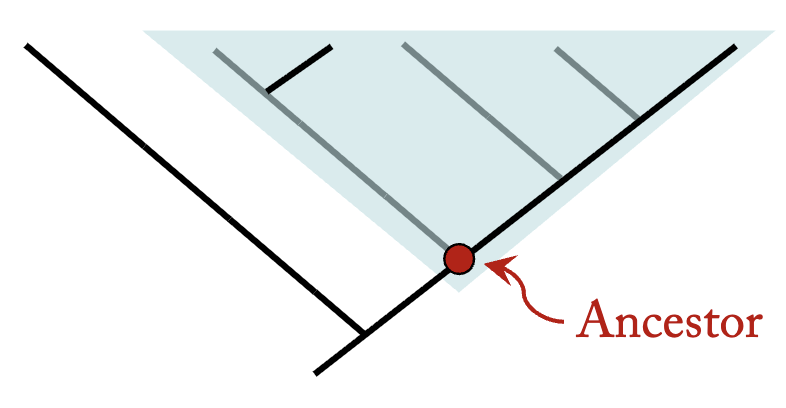
Branch points are called nodes, and represent most recent common ancestors
Branches that share a node are called sister groups
What is the role of the outgroup?
The role of the outgroup is to provide a base to compare the other organisms to, and determine whether the apomorphic state is the loss or gain of a trait.
Apomorphy
Derived evolutionary state (e.g., feathers).
Plesiomorphy
Ancestral evolutionary state (e.g., scales, from which feather evolved).
Synapomorphy
Shared evolutionary innovation, such as feathers shared by birds. The basis of cladograms.
Symplesiomorphy
Shared primitive trait, such as scales among reptiles.
Autapomorphy
Evolutionary innovation present on only one branch on a cladogram.
Homoplasy
A character state that evolved more than once on a cladogram, or evolves and is lost (e.g., character 3).
How can evolutionary relationships be masked by evolutionary change?
Sometimes subsequent change can mask the underlying relationships, fooling parsimony
Ex: A’s unique evolutionary changes masks its true relationships
How can the fossil record help (what do mean when we say the fossil record can be evolution’s time machine)? Given an example.
The fossil record can play a special role, revealing ancestral morphologies before they had become too different to recognize – the fossil record serves as evolution’s time machine
One example of this is Pakicetus, as well as all the intermediate species with diminishing ankle bones being an ancestor of modern whales
How do we use DNA data to build evolutionary trees?
DNA sequences are a wonderful source of phylogenetic information
With parsimony the DNA data matrix is treated exactly like a morphological data matrix
How can parsimony be fooled, that is, what is long-branch attraction? How is it related to statistical inconsistency?
Parsimony can be fooled because the characters of DNA are so simple (the A, T, G, C). Now usually parsimony is OK, but to be sure, it is not used often
This property of increasing data increasing the support for the wrong answer is called statistical inconsistency
How (qualitatively) does Likelihood analysis deal with the problem of long-branch attraction?
For each pair of sequences, the more different they are, the greater the chance that candidate synapomorphies will have arisen by chance. Likelihood analysis weights each candidate synapomorphy in the data matrix by the probability that it was generated by chance given the amount of evolution observed
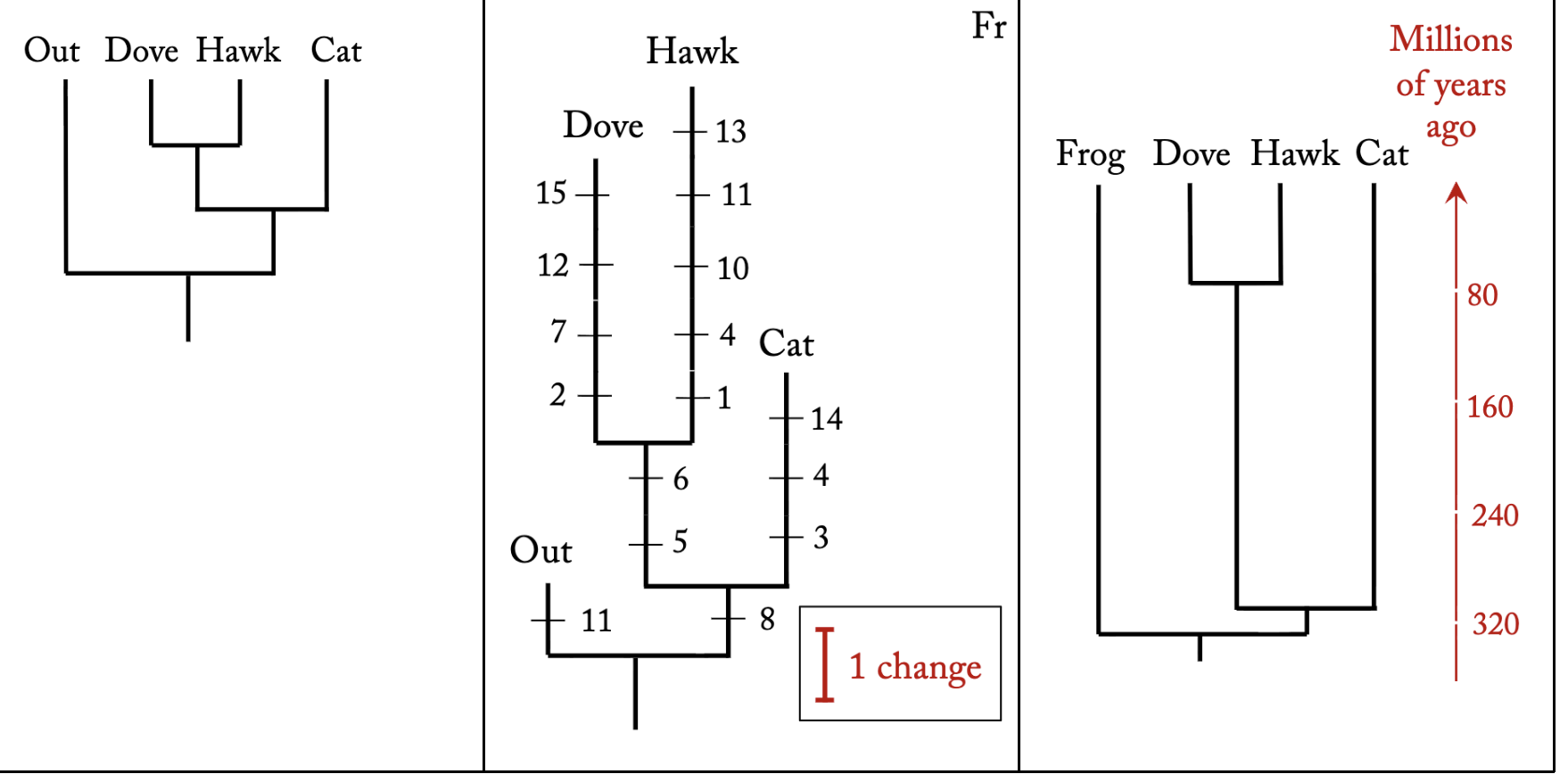
What distinguishes phylograms and phylogenies from cladograms?
Cladogram: just shows relative degree of relatedness (with or without the characters); the branch lengths have no meaning
Phylogram: the branch lengths are proportional to the strength of support (number of changes on the branch)
Phylogeny: the branch lengths are proportional to time
Monophyletic Group
contains an ancestral species and all of its descendants. Monophyletic groups are also called clades.
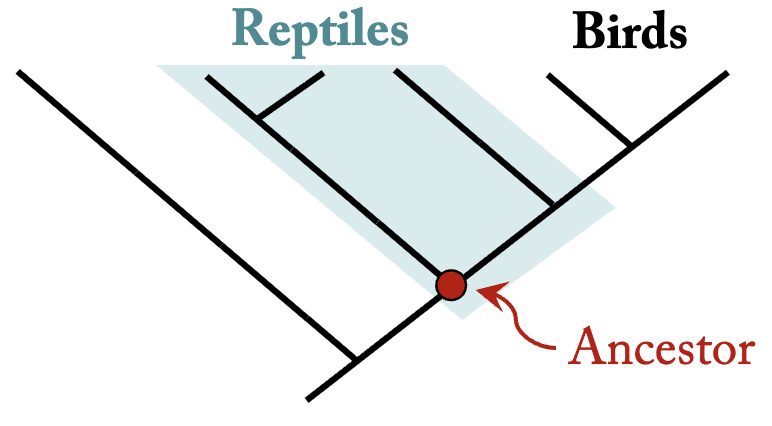
Paraphyletic Group
contains an ancestral species and some but not all of its descendants. Reptiles are paraphyletic because they exclude birds.
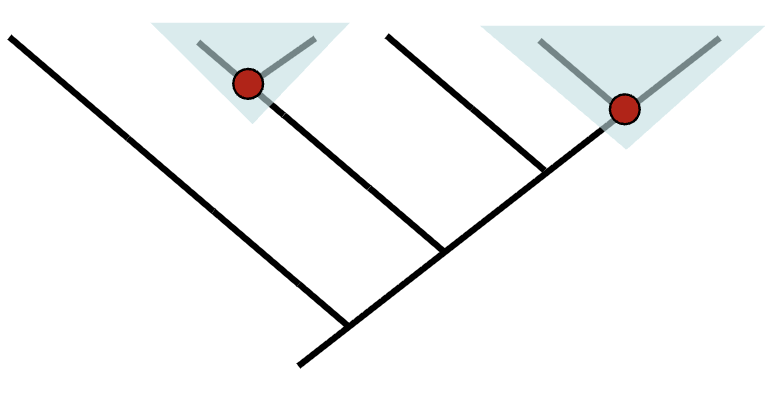
Polyphyletic Group
contains distantly related species but not their most recent common ancestor. Vultures are polyphyletic
Taxonomic groups should be named based on which type of group?
Most systematists feel we should only name monophyletic groups. However, much of our taxonomy predates the building of phylogenies, so paraphyletic groups and their names have become part of our standard vocabulary.
What are the relationships between synapomorphies, homologies, homoplasies, and convergences (parallelisms)?
Homologies are similarities that are signatures of shared ancestry
Synapomorphies are homologs, signatures of shared ancestry
Homoplasies are traits that when viewed in isolation appear to be homologs but on the weight of other evidence may not to be
So, if synapomorphies are equivalent to homologies, then homoplasies are (typically) equivalent to convergences (which are often also called parallelisms, although some use the terms slightly differently)
Can you give some examples of convergences?
stem succulence, plant morphology, nectar feeding in birds, etc.
What is the Phylogenetic Species Concept?
A species is the smallest monophyletic group (lineage) distinguishable by a unique set of typically genetic (but can be morphological) traits, that is, synapomorphies).
Why might the Phylogenetic Species Concept lead to the recognition of more species than the Biological and Morphological Species Concepts
For all species concepts, there will be a time lag between the allopatric event and when the populations will be called different species, the time it takes to gain enough differences to become morphologically distinct, or to no long interbreed.
It is quite possible that two monophyletic groups will be able to interbreed, but simply haven’t for a while
Thus, the Phylogenetic Species Concept is likely to lead to the recognition of more species than would be recognized by the Biological and Morphological Species Concepts
[Exit Ticket] What is the relationship between homology, synapomorphy and homoplasy?
Synapomorphies are homologs, for example the limbs of tetrapods. Homoplasies are characters that in isolation we might have thought were homologs but turn out not be (typically representing convergences).
How were phylogenies used to make better flu vaccines?
The fastest evolving viruses tend to seed the following year’s infections. Why? Our immune systems selects for them, as they are the ones most likely to find a way of evading our attempt to kill them. So, the CDC now picks the longest branch viruses for the next vaccine
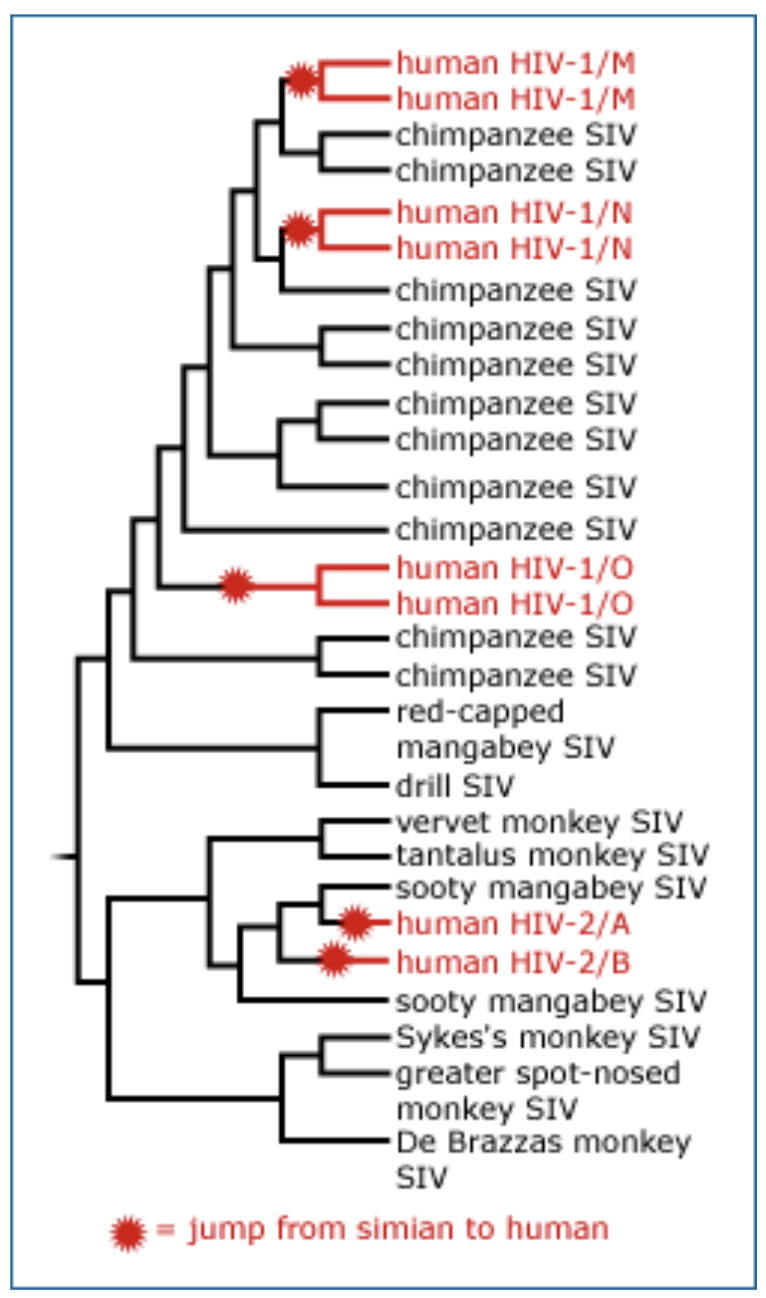
How were phylogenies used to determine where the HIV virus came from, and how many times it crossed over into humans?
HIV (Human immunodeficiency virus) is closely related to SIV (simian immunodeficiency virus)
A phylogeny of human and primate sequences shows that the origin of the human virus is polyphyletic, it has many separate origins!
Tree of human HIV sequences, showing multiple origins from ‘bush meat’ (chimps, gorilla, mangebey)
Phylogenies also track the timing and geographic paths of HIV infection in the US (it came via the Caribbean ...)
A “molecular clock” was used, where the number of DNA changes was used to infer the amount of time elapsed, allowing us to date nodes on the tree.
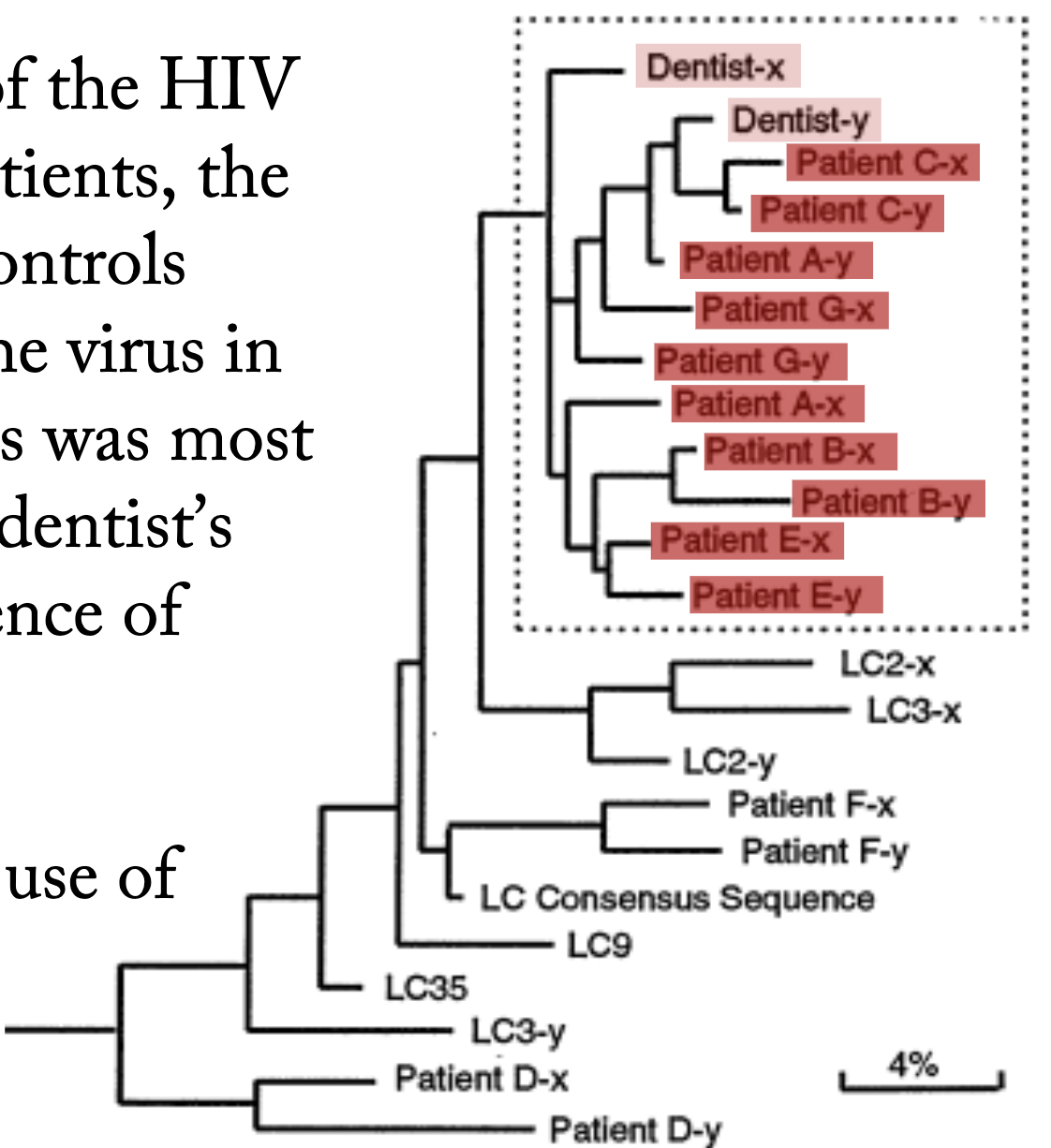
How were phylogenies used to determine whether a dentist infected his patients with HIV?
A phylogenetic tree of the HIV sequences of these patients, the dentist, and several controls (LCXs) shows that the virus in some of these patients was most closely related to the dentist’s virus, providing evidence of transmission
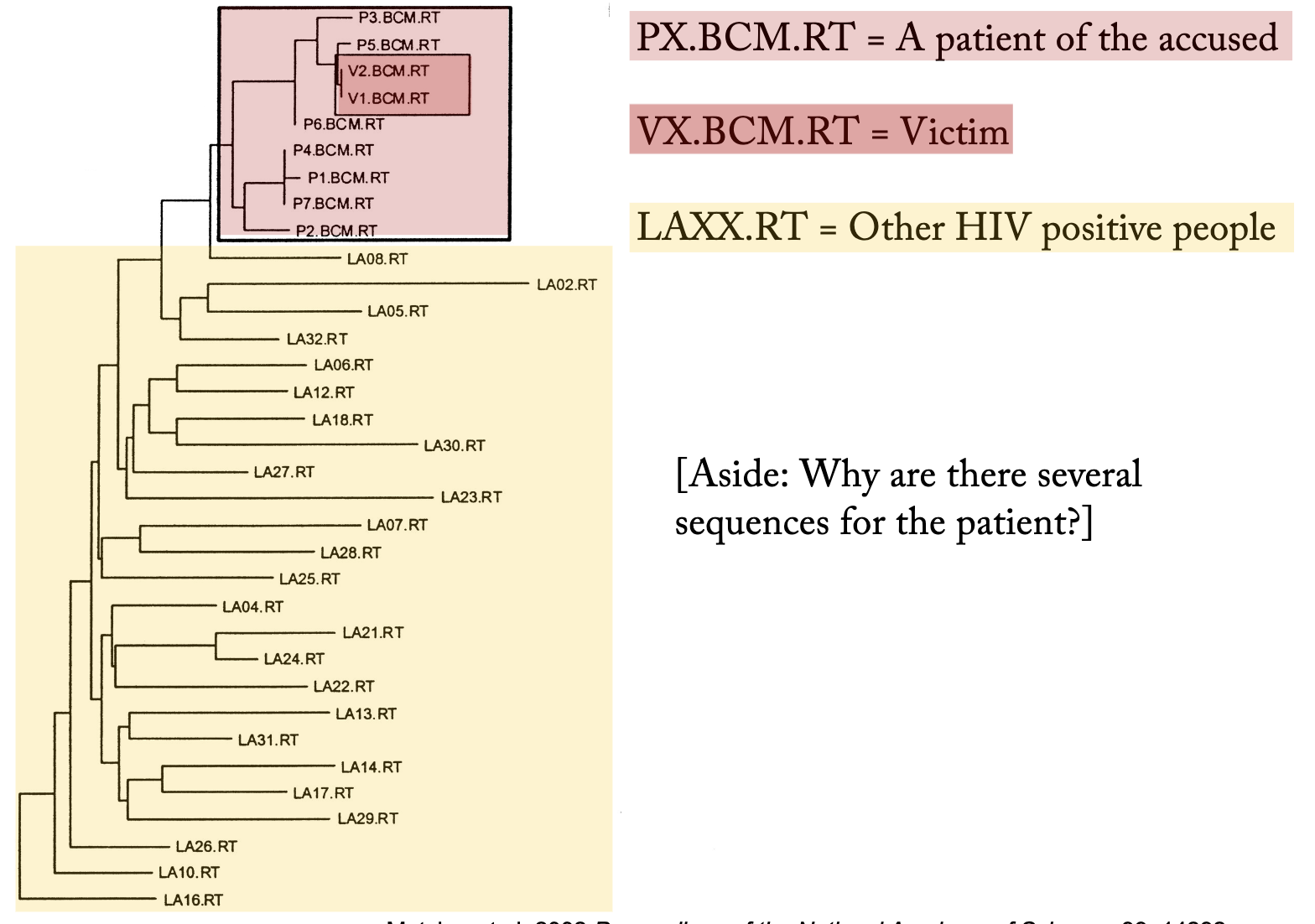
How were phylogenies used in a law case where a doctor was accused of injecting his girlfriend with HIV?
Phylogenetic analyses of HIV-1 sequences were used as evidence, the first use of phylogenetic analyses in a criminal court case in the United States
Analysis of DNA sequences from the victim, the patient, and some local HIV-1-positive people showed the victim’s HIV-1 sequences nested within a lineage comprised of the patient’s HIV-1 sequences – the paraphyly of the patient’s sequences implied transmission from the patient to the victim.
What was the hypothesis of the molecular clock?
Until the mid-1960s the only way to estimate times of divergence between lineages was to use the ages of oldest fossils
And then came the Molecular Clock, the hope that differences in the DNA between species accumulate at a universal (stochastically) constant rate, and thus that the DNA differences between living species could be used to date their times of divergence (analogous to radioactive decay)
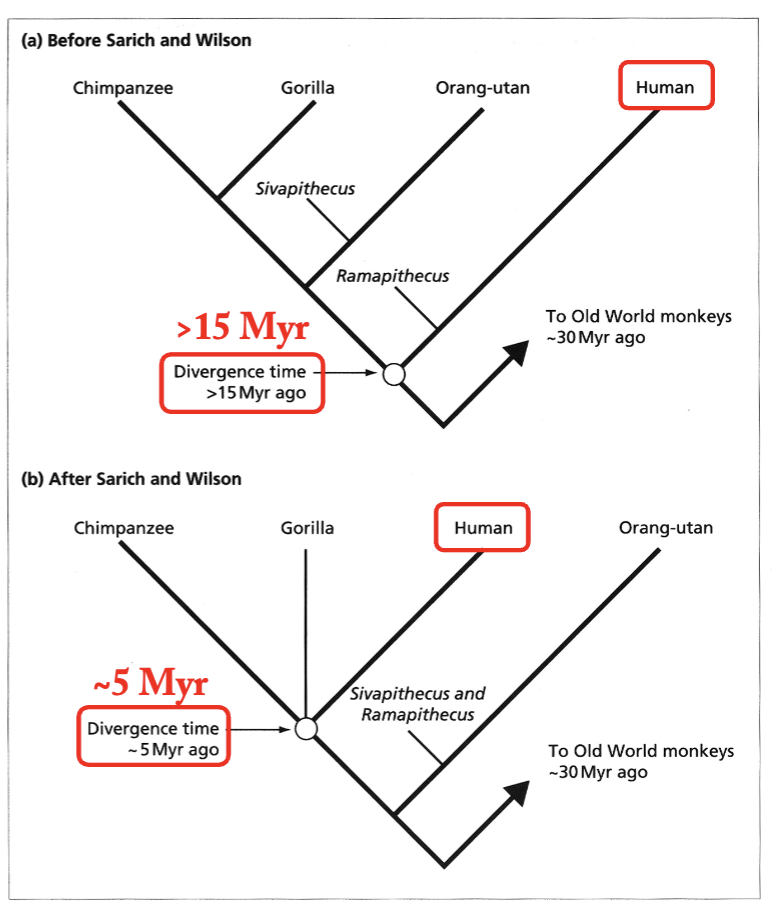
How did the molecular clock change our view of human origins?
Traditional view of our relationship to the Great Apes – as far away as possible!
Using an estimated divergence between Homo sapiens and the Old-World Monkeys of 30 myr, they concluded that Homo and Pan (chimps) were (1) sister groups and (2) only diverged 4-5 million years ago
Is there a universal rate of DNA change? If not, why not?
There is no universal clock, even though the rate of change is relatively constant within individual clades for neutrally evolving DNA sequences.
Neutral rates of evolution:
– What does this mean?
Neutral changes are those that are neutral with respect to selection, i.e., not subject to selection.
The Genetic Code: synonymous changes don’t change the amino acid; non-synonymous changes do
Within genic regions of DNA, synonymous changes are (essentially) neutral.
What is the difference between synonymous and non-synonymous mutations?
The Genetic Code: synonymous changes don’t change the amino acid; non-synonymous changes do
What is the difference between a mutation and a substitution?
Mutations are changes in the DNA
Substitutions are mutations that become fixed in a population
Mutation Rate (μ) = Rate of un-repaired DNA changes per nucleotide position per year per individual.
Substitution Rate = Rate that mutations become fixed in the population. Once fixed, a substitution becomes a characteristic (synapomorphy) of that population
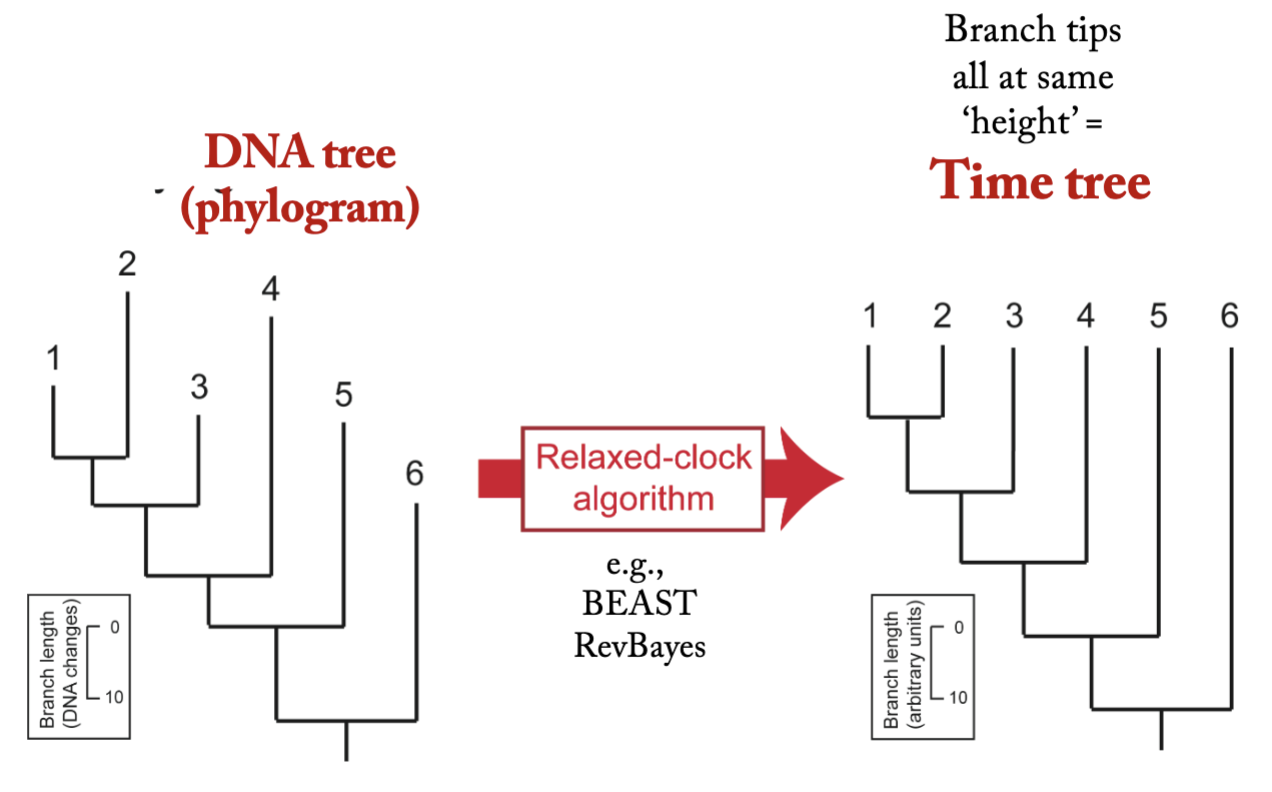
Can we use DNA distances to estimate divergence times even if there is no universal clock?
– What technique is used to do this?
Now we are in a post-molecular-clock era. Rather than assume an average rate of change, we take our phylogram (the cladogram with the branch lengths measured in DNA changes) and ‘rate smooth’ it, to convert it into a phylogeny (also called a timetree or a chronogram; I will not explain how).
Rate Smoothing (also called a relaxed molecular clock analysis) converts a phylogram into a time tree
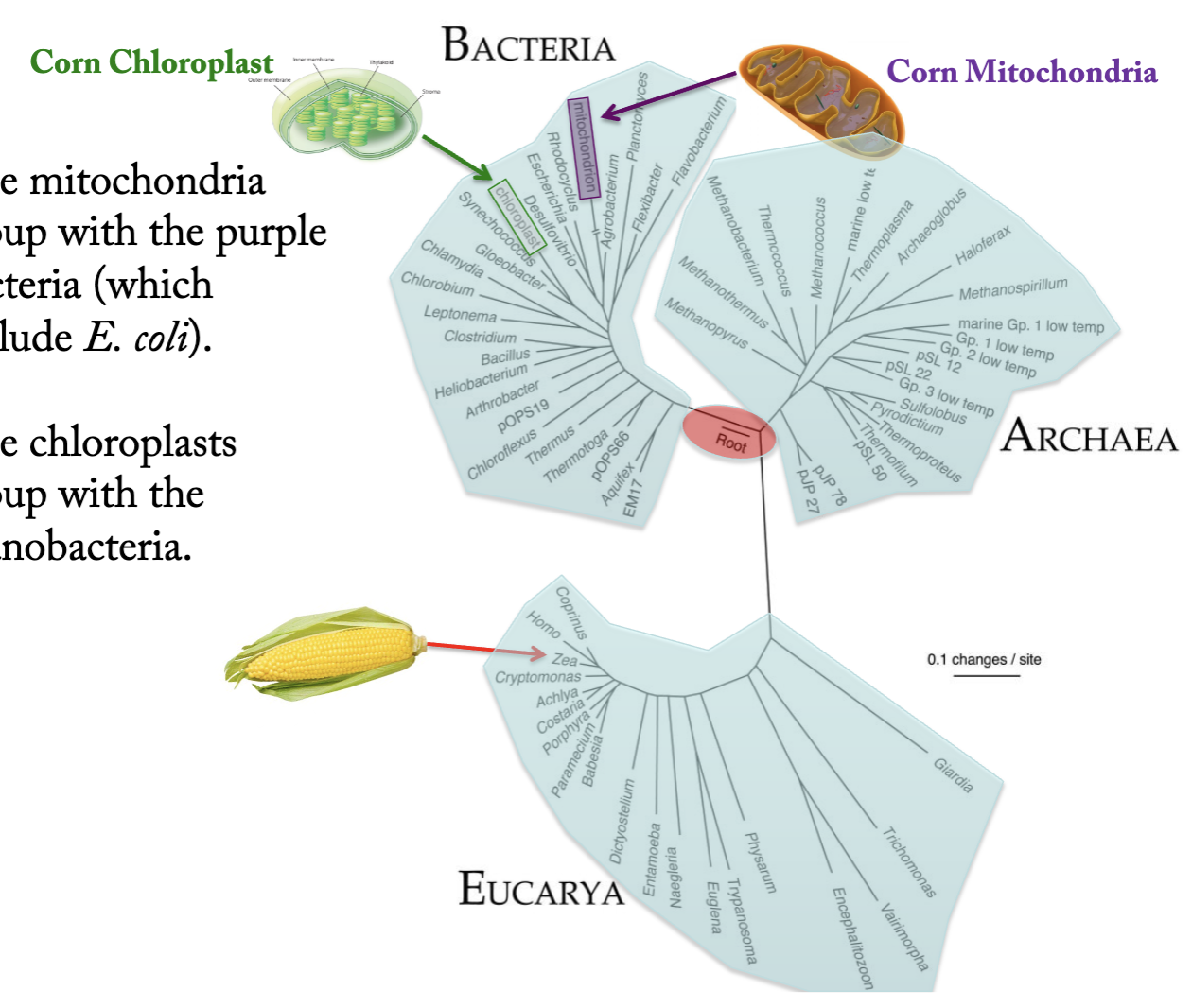
• Why has the SSU rRNA been such a valuable phylogenetic tool?
– What did it reveal about the origin of mitochondria & chloroplasts?
The Small Subunit ribosomal RNA (SSU rRNA): A Universal Phylogenetic Tool
Part of the machinery that makes protein.
All organisms have it.
Very conserved, so can we can recognize it in all species
Revealed 3 Great Domains in the Tree of Life: Bacteria, Archaea, Eucarya
The mitochondria group with the purple bacteria (which include E. coli).
The chloroplasts group with the cyanobacteria
DNA tree shows that mitochondria and chloroplast are derived from prokaryotic bacteria