Gene cloning and manipulation
1/47
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
Taq polymerase
72-75C optimum
No proofreading
Requirements of primers
20-30bp for specific priming
C/G clamp to prevent drop off
Roughly same temperature for annealing step
Avoid complementarity to prevent primer dimers
Avoid secondary structure (e.g. hairpin loop) formation
Methods of improving primer specificity
Decrease concentration Mg2+ to decrease mispriming (Mg2+ offsets -ve charge on phosphate backbones, reducing repulsion)
Touch down PCR
Hot start PCR
Nested PCR

Describe touch-down PCR and draw a graph
Start with highest temperatures, where all will be bound
Decreasing temperature every cycle
First successful reactions will be under the strictest conditions
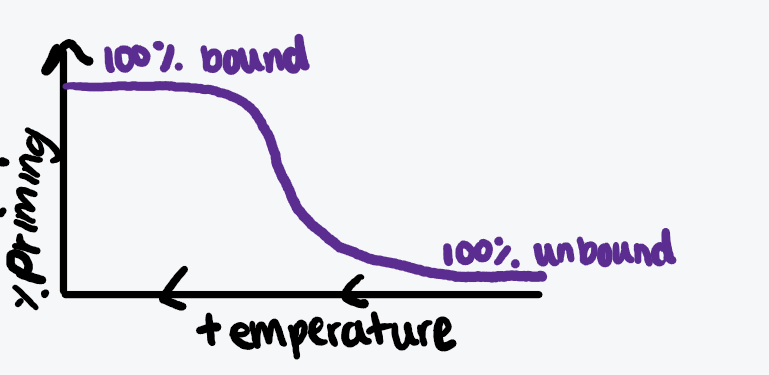
Describe hot-start PCR
Polymerase is made to be completely inactive until annealing temperature is met
Can either add polymerase manually when temperature is reached, or use JumpStart Taq which is inactive below 70C
Describe nested PCR and draw a diagram
In round 1, primers are sometimes non-specific and can bind in multiple places
In round 2, a nested primer is used that will bind inside the target sequence, but not inside the misprimed sequence
Used when round 2 primer could not be used in round 1 as too non-specific, but once sequence is narrowed down, it can be used

What is the multiple cloning site
Series of restriction sequences that will be recognised by various restriction enzymes
Where is the MCS in pBluescript and why is this useful
Within the functioning LacZ’ gene
DNA insertion will inactivate the gene, allowing for blue(non-insert)/white(insert) selection
What is the LacZ’ gene?
Encodes beta-galactosidase
When combined with the rest of an E.coli genome, will cause colonies to appear blue when exposed to IPTG and X-gal
T7 phage promoter
Increases replication rate
Purpose of ampicillin resistance gene in a plasmid
Can kill all cells that didn’t take up the plasmid using ampicillin
Histags
Helps purifiy proteins
Follow with thrombin site to cleave tag once purifiedW
What are good characteristics of cloning plasmids
MCS
LacZ’
Ori site
T7 phage promoter
Ampicillin resistance
Histag
Thrombin site
OriC site
Promotes plasmid replication inside the cell
What is favourable when choosing an E.coli strain to transform into
High efficiency of transformation
Lack of DNA-degrading enzymes
Recombination deficiency to maintain plasmid stability
Blunt cloning
PCR usually makes blunt ends
Treat plasmid with blunt-end RE (e.g. EcoRV)
DNA ligase
Non-directional
TA cloning
Taq adds a non-templated A to the end of PCR products
Clone using a different high-fidelity polymerase
Use taq for final cycle, will add an A
Clone into commercially available vector that has a single T overhang
Efficient
Non-directional
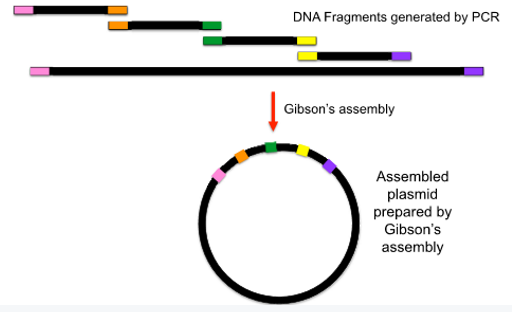
Gibson’s assembly, draw a diagram
Joins multiple fragments directionally, regardless of sequence
Use in-silico primer design to create a series of fragments with overlapping ends
Treat with T5 exonuclease that causes 3’ overhanging sticky ends
Annealing with polymerase will seal the nicks
Golden gate cloning
Simultaneous and directional assembly of multiple DNA fragments using type IIS restriciton enzymes and T4 DNA ligase
The RE cleaves DNA just after recognition sequences so recognition sites are removed from the insert
RT/qPCR
Before first PCR replication, add reverse transcriptase at a normal temperature
Will turn all mRNA into DNA
Amount of DNA produced during a known number of PCR cycles correlated with the amount of mRNA started with
SYBR green dye and qPCR
Used in qPCR
Binds to dsDNA and fluoresces
Intensity of fluorescence above background level is measured
Number of PCR cycles to generate fluorescence above a threshold value is the CT
Sequence-specific fluorescent probes and qPCR
Fluorophore at 5’ end, quencher at 3’ end
Probe binds to DNA downstream of primer, no fluorecence
5’ → 3’ exonuclease activity of DNA pol will cleave and release the fluorophore
Fluorescence is now detectable
Purification of his-tagged proteins
Immobilize on columns lined with nickel ions
Wash away unbound
If cleavage site present: add protease to release protein from column
If lacking cleavage site: elute by imidazole that competes for cation sites and displaces fusion protein
Considerations when expressing proteins in E.Coli
Doesn’t use splicing, so unless cDNA is used, introns will be expressed
Protein may not be easy to isolate without loss of biological activity
Protein may be lysed or secreted
Could be toxic
Describe immunoprecipitation as a way of identifying interacting proteins
Antibody fixed to protein A/G beads that binds specifically to protein of interest
Verify that antibody only recognises that protein and none others in that mixture
Magnets can capture protein A/G beads, bringing out the protein of interest and also any proteins that are attached to it
Mass spec to determine interacting proteins
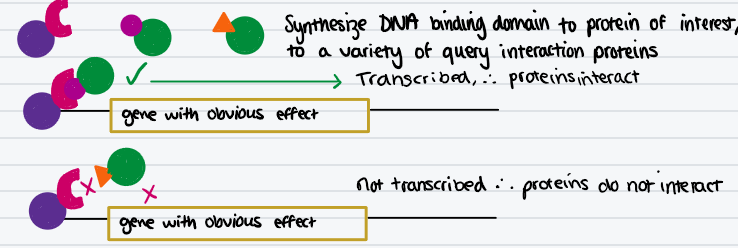
Describe the yeast two-hybrid system for identifying interacting proteins, draw a diagram
S.cerevisiae transcription factor GAL4 has a DNA binding domain and an activation domain. When joined together, activate transcription
Joining doesn’t have to be direct - can be activated by bridging with proteins
Synthesize DNA binding domain to protein of interest, and activation domain to a variety of query interaction proteins
See which combinations cause transcription
Mutagenesis PCR, draw a diagram
Mutant olignoucleotides that are entirely complimentary except for the desired point mutation
Transform into E.coli
Will either replicate (introducing the point mutation on both strands) or repair

Simply, what are three ways of reducing protein expression?
RNA interference
CRISPR/Cas9
Gene inactivation by homologous recombination
Reducing protein expression with RNA interference
Post transcriptional reduction
Long dsRNA processed into siRNA by RNAase III (Dicer)
Made of one guide strand and one passenger strand (that will be destroyed)
siRNAs assemble with Ago2 to form RISC
Perfect complementarity
Duplex formation causes endonucleolytic cleavage by Ago2
Remaining mRNA degraded in general cellular decay pathways
What determines which strand is the guide and which is the passenger in siRNAs
The stability of each 5’ end
When can and can’t siRNA be used in humans
Can’t when need to introduce long dsRNA as triggers an interferon response (mistaking for virus e.g. rotavirus)
Use synthetic shRNA to transfect, which will then be processed like miRNAs to give rise to the guide strand that still targets mRNA without an interferon response
Describe the components of CRISPR/Cas9 used in gene editing
Cas9 dsDNA nuclease
gRNA that combines crRNA and tracrRNA (specific to the sequence being edited, generated by cloning of oligonucleotides specific to the target DNA)
How does CRISPR/Cas9 edit genes
gRNA binds with perfect complementarity
Cas9 causes ds break that triggers repair mechanisms
Repairing is good but not great, so can make error
If repair is perfect, gRNA binds again and Cas9 cuts
Repeats until microinsertion or deletion is complete
What do big deletions using CRISPR/Cas9 editing require?
Two gRNAs for two ds breaks that ligate together to completely remove that section of DNA
How can dead Cas9 be used in gene editing?
Blocks transcription
Can be bound to other enzymes or fluorophores
Method of specific recruitment on the genome
Gene inactivation by homologous recombination
Knock out of endogenous genes by swapping them with a mutant gene
Insert a selectable marker into a clone of the gene desired to be disrupted
Introduce into non-replicating DNA
Select for maker, only way for cell to survive is for it to replace the existing gene in the genome with the disrupted marker gene
Endogenous gene is now replaced by disrupted copy
Analysing protein function using microinjection of transgenic animals
Microinject DNA into pronucleus of fertilized eggs
Random integration
Transplant to psuedopregnant mother
10-20% offspring will develop into the transgenic organism
Analyzing protein function using cultured embryonic stem cells and transgenic animals
Undifferentiated cells can be manipulated
Reinjected into developing embryo
Identify suitable cell lines that have integrated the DNA using PCR
Develop
Form a mosaic animal
Can be mated to WT to achieve heterozygous mutant and if mutation is dominant, will be completely presenting the mutation
Use of bromophenol blue in gels
Visualise loading
Can monitor how long the gel has been running
Advantage of using two different restriction enzymes to clone a DNA fragment into a plasmid
Directional
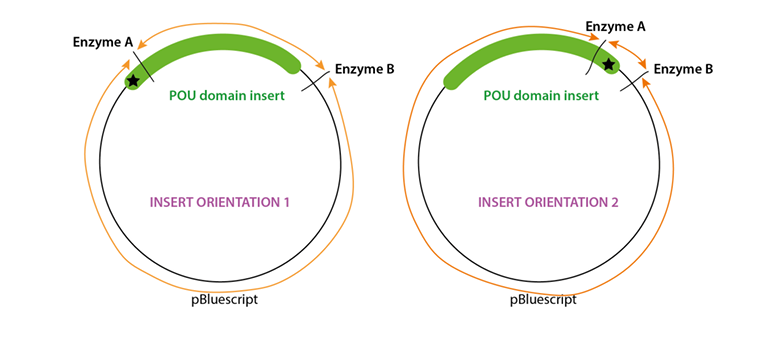
How to check insert orientation without sequencing, draw a diagram
Restriction digest using two enzymes that cut asymmetrically within the insert
Analyse using agarose electrophoresis alongside molecular weight markers
Reasons why PCR has failed, besides forgetting to add any components
DNA template quality (degraded or contaminated)
Primers have sequences that form primer dimers or secondary structures e.g. hairpins
Incorrect annealing temperature
Too little Mg2+, too much primer-template repulsion
Why could a variety of sizes of DNA bands be obtained after a PCR when only one is expected? How to resolve?
Non-specific primer binding
Increase annealing temperature
Decrease Mg2+ concentration
Change primers
How much DNA is required for LCN PCR?
100-200pg of cells - roughly one billionth the mass of a grain of rice
What are advantages of using a reporter gene in an assay for expression
Analyse promoter function
Can easy be visualised (GFP) / assayed (beta-galactosidase) to interpret expression from a particular promoter
Analyse other regulatory elements or sequences (enhancers, miRNA binding sites)
Can compare promoter function
Study localisation
Describe how DNA footprinting is carried out
DNA is one end-labelled using 32P
Protein incubated with dsDNA
Mildly digested with DNAase, each DNA molecule is cut once
Separate on agarose
Visualise radioactivity
DNA without protein will have a continuous fragment ladder (cuts at a random points on every strand)
DNA and protein will have a gap (footprint) where DNA has been protected and remains associated with protein
Where are hydrophobic amino acid interactions most important in a protein?
Core
Stabilize structure
What are charged side chains of amino acids especially good at doing?
Stabilizing helix dipole of alpha helices