BSCI 1510 Test 3
1/166
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
167 Terms
What must cells do during the cell division cycle?
-Grow to double in size
-Duplicate all macromolecules
-Repair any damage to DNA
-Segregate chromosomes
-Divide into two cells
M Phase
Consists of mitosis and cytokinesis
Interphase
Consists of G1 S and G2 in which a cell continues to transcribe genes, synthesize proteins, and grow in mass
Mitosis
Segregation of chromosomes
Cytokinesis
Cytoplasm of two cells divide
G1
cell growth
S phase
DNA replication
G2
DNA repair/ ensure completion of DNA replication
Cell Cycle Control System
Regulatory proteins that make sure cells replicate all their DNA and organelles and divide in an orderly manner.
-Also makes sure that the events of the cell cycle such as DNA replication, mitosis and so on occur in a set sequence and that each process has been completed before the next one begins
Late G1 checkpoint
Control system confirms that environment is favorable for DNA replication (sufficient nutrients and specific signal molecules in the extracellular environment
If environment is not favorable the cell is sent back to G0
Point in the cell cycle where the control system is regulated by outside influences
Sometimes referred to as “Start” since in this late G1 checkpoint cells commit to begin and complete a full cell cycle
Late G2 checkpoint
Confirms DNA is undamaged and fully replicated
Ensures cell does not enter mitosis unless DNA is fully in tact
Midway through mitosis checkpoint
Makes sure duplicated chromosomes are attached to spindle fibers before the spindle pulls them apart and segregates them into two daughter cells.
What is progression through the cell cycle dictated by?
Cyclins and cyclin dependent kinases
How does cyclin work?
Cyclin regulatory proteins attach to cyclin dependent kinases to phosphorylate proteins at different transition points in the cell cycle
Cyclin-Cdk complex
Phosphorylate key proteins in the cell that are required to initiate particular steps in the cell cycle.
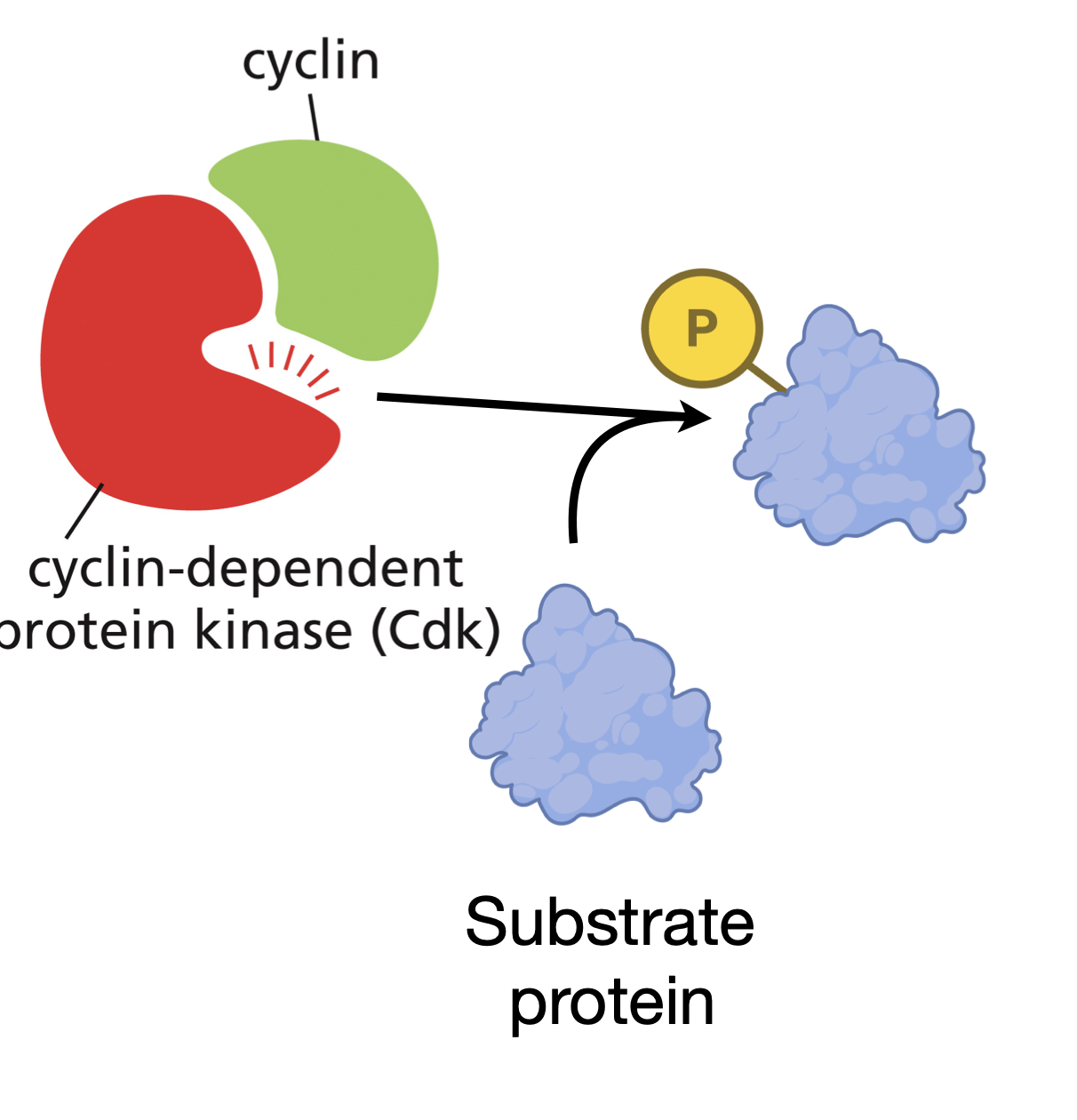
Different types of cyclin
G1/S cyclin CDK complex is activated midway through G1 phase
S cyclin CDK complex activated right before S phase
M cyclin CDK Complex right before commencing M phase
Cyclins themselves have
no enzymatic activity but must bind to CDKs in order for the kinases to become active.
Concentration of Cyclin vs CDK
The concentration of cyclin varies throughout the cell cycle but the concentration of CDK remains the same however the activity of CDK cycles.
G1-Cdk complex is made up of
Cyclin D and Cdk 4, Cdk 6
G1/S Cdk complex is made up of
Cyclin E and Cdk 2
S Cdk complex is made up of
Cyclin A and CDK 2
M Cdk complex is made up of
Cyclin B and CDK 1
What helps drive the assembly and activation of cyclin CDK complexes?
Changes in the concentration of cyclin
How does cyclin destruction help transition a cell from one phase of the cell cycle to the next?
Like cyclin accumulation, cyclin destruction can help transition from one phase of the cell cycle to the next. For example, M cyclin degradation which leads to the inactivation of M Cdk leads to the molecular events that take a cell out of mitosis.
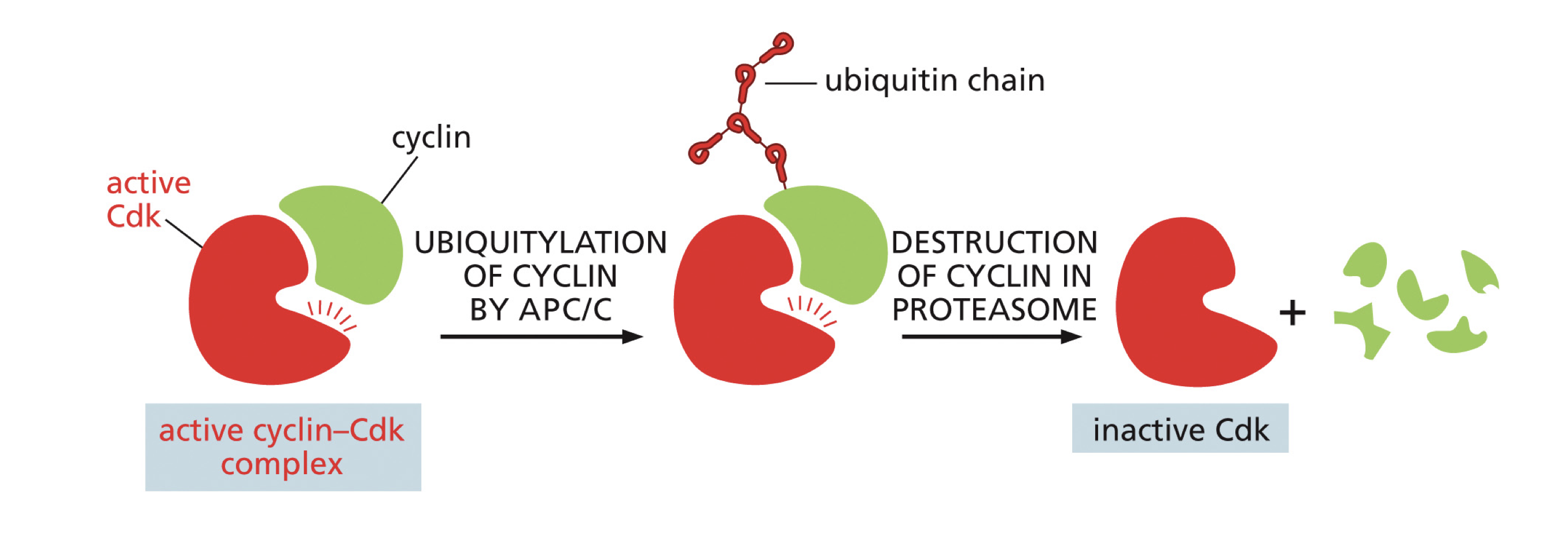
Protein Degradation
For cyclins to inactivate the cyclin CDK complex. Occurs when a ubiquitin chain is attached to cyclin by APC/C then cyclin gets sent to a proteasome to get degraded. Once cyclin is degraded the CDK becomes inactive.
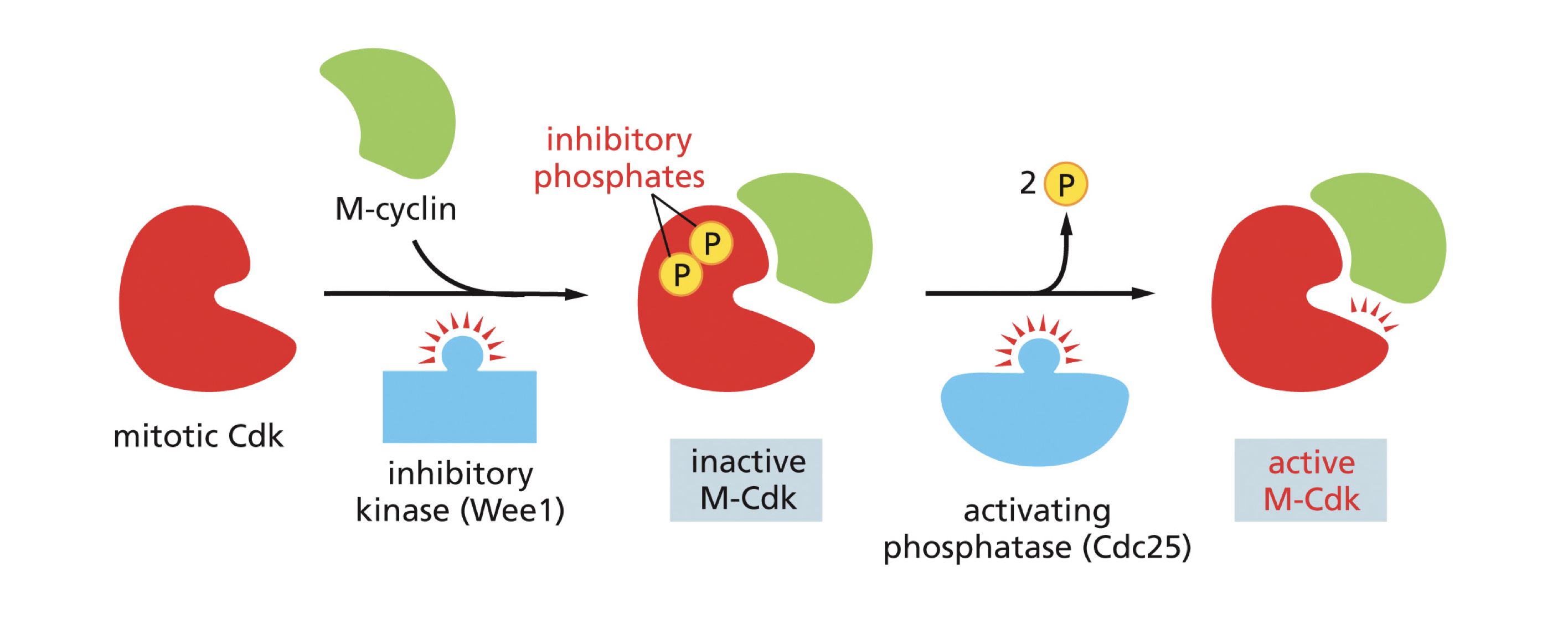
Inhibitory Phosphorylation
Cyclin CDK complexes contain inhibitory phosphates that are bestowed to them by an inhibitory kinase named Wee 1 and in order for them to become active a phosphatase named (cdc 25) must remove these inhibitory kinases
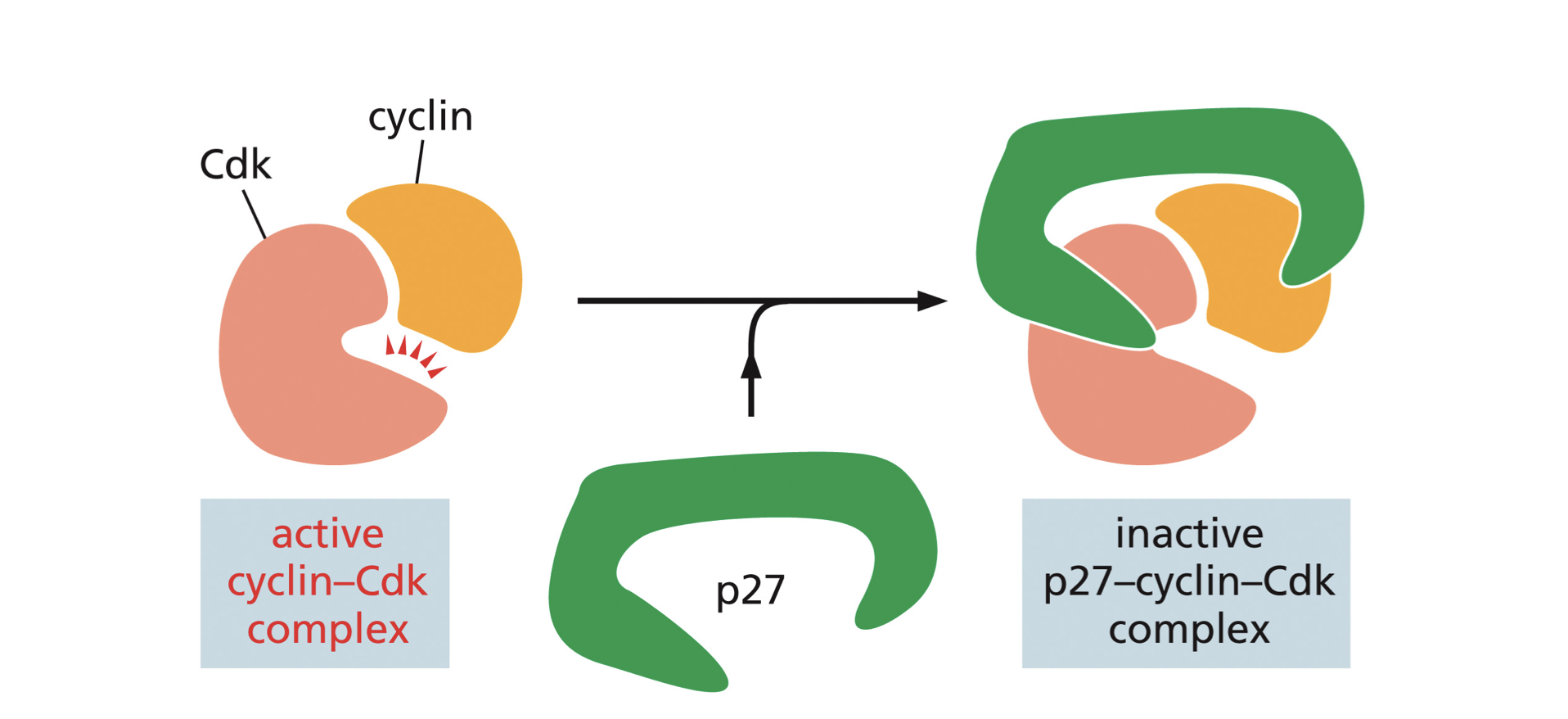
Blocked by Inhibitory proteins: cyclins and CDKS
Proteins such as P27 bind to an activated cyclin-CDK complex which prevents the CDK from phosphorylating target proteins for progress from one phase to another
Full G1 phase
-Enter Cell Cycle or exit cycle (G0/quiescence)
-Depends on intrinsic signals (cell size)
-Depends on extrinsic signals (nutrients/mitogens)
-Requires Cyclin D complexed with CDK 4 or CDK 6
Full S phase
-Genome duplication
Once and only once per cell cycle
All or Nothing
-Requires S CDK activity; Cyclin E or Cyclin A complexed with CDK 2
-Centrioles are duplicated
Full G2 phase
-Ensures that DNA replication is complete
-Repair/resolve any DNA damage prior to mitosis
M phase
-Chromosome Segregation
-Requires M-CDK- Cyclin B/CDK 1
What provides the pulling force of mitosis?
Microtubules provide the pulling force of mitosis since the plus end of the microtubule combines with connecting protein complex to attach to the kinetochore of chromosomes to pull them apart.
Five phases of Mitosis
Prophase
Prometaphase
Metaphase
Anaphase
Telophase
Prophase
Duplicated chromosomes condense
Mitotic spindle assembles
Centrosomes move apart
Prometaphase
Breakdown of nuclear envelope
Chromosomes attach to mitotic spindle through kinetochore
Active movement of chromosomes
Metaphase
Chromosomes align at the equator of the spindle
Each sister chromatid is attached to opposite poles of the spindle
Anaphase
Sister chromatids pulled to opposite ends of spindle pole
Kinetochore microtubules get shorter
Spindles move outwards
Telophase
Chromosomes arrive at spindle poles
Nuclear envelope reassembles around each set of chromosomes
Chromosomes begin to decondense
Cytokinesis
Cytoplasm is divided in two by contractile ring
Two daughter cells are formed each with their own respective nucleus
Retinoblastoma
Rare cancer in young children caused by mutation in the RB1 gene (tumor suppressor). RB is a target of Cyclin D and CDK4,6 complex and prevents entry in the S phase
Early Experiments by Griffith on bacterial transformation
When pathogenic bacteria that was heat killed (no longer infectious) was injected into a mouse along with live harmeless bacteria this combination proved unexpectedly lethal: not only did the animals die of pneumonia but Griffith found that their blood was teaming with live bacteria of the pathogenic form. The heat killed had somehow converted harmless bacteria into the lethal form. This provided evidence that genes are made up of DNA
Avery, Macleod, and McCarty (1944)
Demonstrated that DNA is the genetic material that led to the results of Griffith’s paper.
They basically purified RNA, protein, lipid, carbohydrate, and DNA and then incubated the harmless R strain which each of these purified forms
Only the DNA fraction was able to convert the harmless R strain into the lethal S strain
Which experiment showed indefinitely that genes are made up of DNA?
Hershey and Chase
What did Hershey and Chase do in their experiment?
Hershey and Chase radioactively labeled the proteins in the viruses with Sulfur-35 and the DNA with phosphorous 32. They then allowed the labeled viruses to infect E coli and the mixture was then disrupted by a brief pulsing in a warring blender and centrifuged to separate bacteria from empty viral heads. When researchers measured the radioactivity they found that much of the phosphorous 32 had entered the bacterial cells while the vast majority of the sulfur 35 labeled proteins remained in the solution with the spent viral particles
What did this show?
Since the Phosphorous 32 was found inside the bacterial cell and made its way into subsequent generations of virus particles confirming that DNA is heritable genetic material
Watson and Crick
The molecular structure of DNA was solved 1953 by Watson and Crick who used Xray data obtained by Franklin and Wilkins.
What do nucleotides consist of?
A base (purine or pyrimidines), a phosphate, and a sugar (ribose or deoxyribose).
Where are bases and phosphates linked on nucleotides?
Bases are linked to Carbon 1’ of the sugar while the phosphate is linked to Carbon 5’ of the sugar
What is the difference in the nucleotides of RNA and DNA?
Carbon 2’ has a hydroxyl group for RNA while Carbon 2’ has a single Hydrogen group for DNA
Where are purines and pyrimidines linked to on the pentose sugar?
Purines are linked to the N1 position of the pentose sugar while Pyrimidines are linked to the N9 position of the pentose sugar.
Purines
Adenine and Guanine
Pyrimidines
Cytosine
Thymine (DNA only)
Uracil (RNA only)
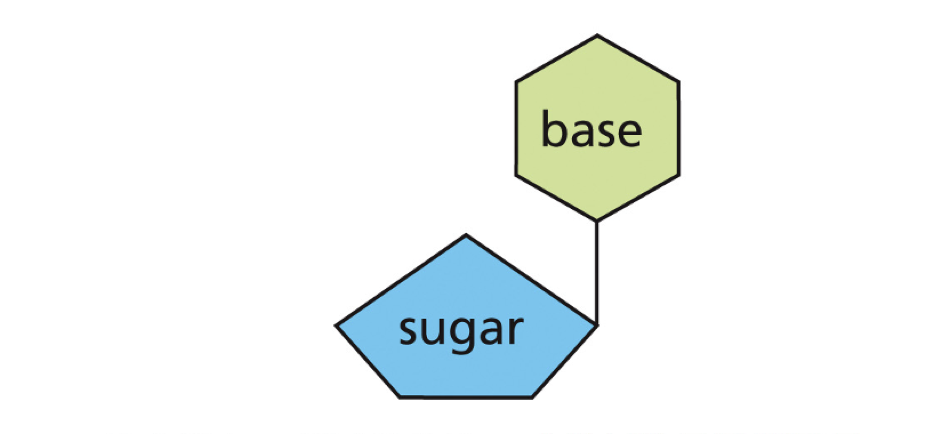
Nucleoside
BASE+SUGAR
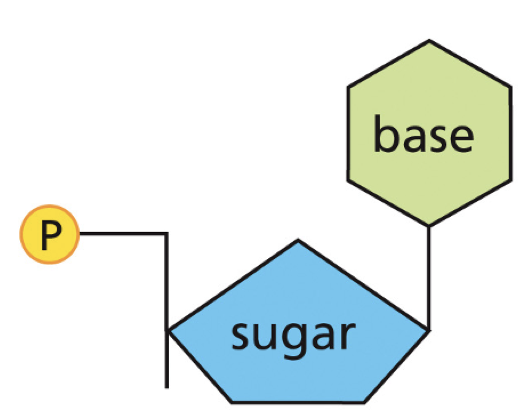
Nucleotide
BASE+SUGAR+PHOSPHATE
How are nucleotides linked together?
Through phosphodiester bonds to form nucleic acids
How do nucleic acids establish polarity?
By having a 5’ end and a 3’ end
5’ end contains
a phosphate
3’ end contains
Contains hydroxyl (-OH) group
The DNA helix contains
two antiparallel strands of DNA with the bases connecting the two strands (bases connect through hydrogen bonds).
Pitch
One full helical turn of DNA which is 10.5 base pairs
How are bases fully hydrogen bonded both as single stranded DNA and double stranded DNA?
Bases are fully hydrogen bonded in a single strand with water and in a double strand with other bases
B Form DNA
The most common biological form; A right handed helix meaning the strands are tilted right when crossing the front of the helical axis

B DNA
has 10.5 base pairs per turn (pitch)
rise=3.4 angstroms per base pair (van der waals thickness)
Pitch= 34 angstroms
Helix Twist= 36* per base pair
Helix diameter= 20 Angstroms
CG base pairs have
3 hydrogen bonds
AT base pairs have
2 hydrogen bonds
Chargaff’s rule
%A=%T
%C=%G
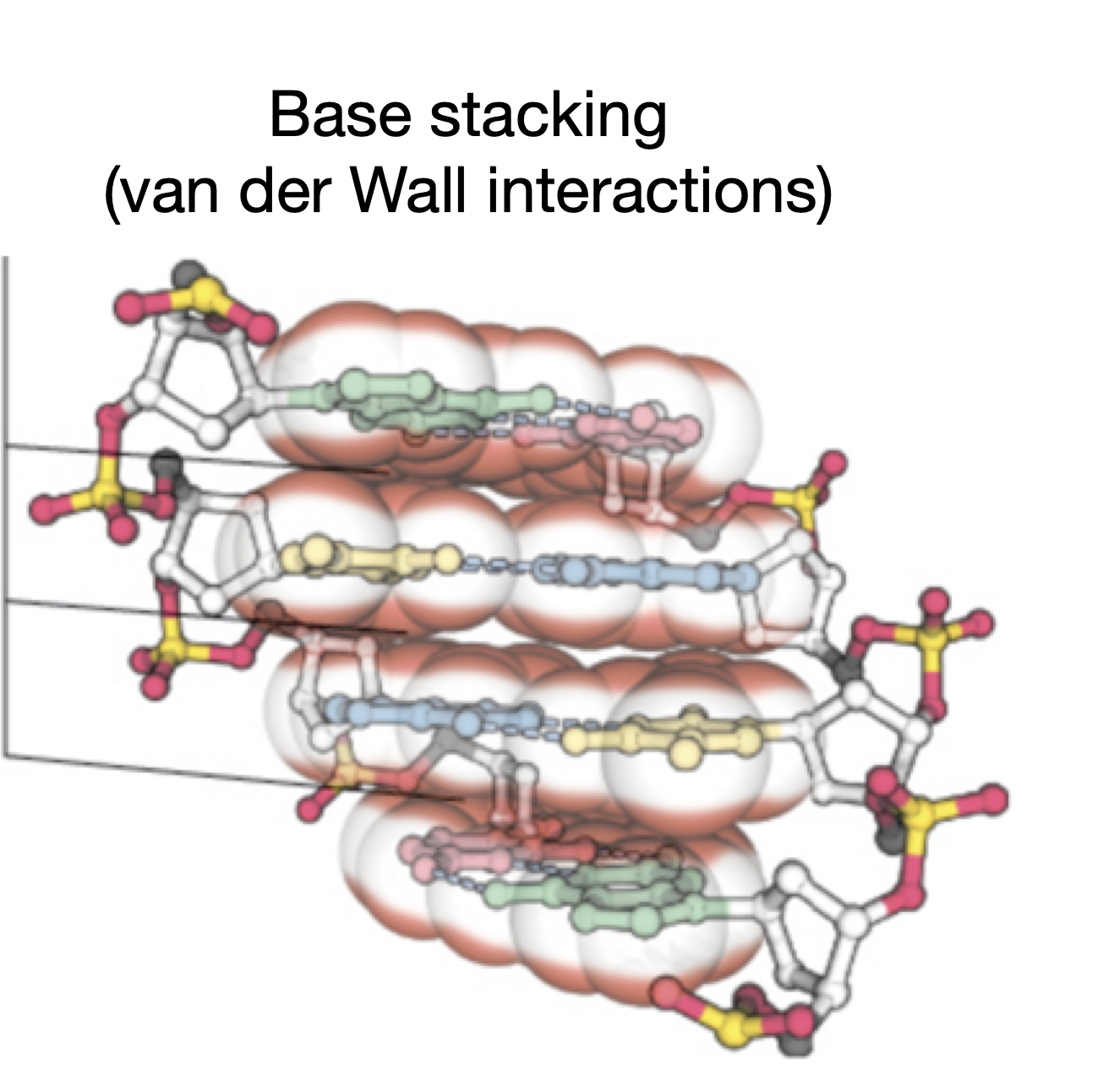
What are the most dominant force in stabilizing the double helix?
Base stacking interactions
C value Paradox
Some simple organisms have much larger genomes than more complex ones because most DNA does not code for proteins-much of it consists of noncoding regions such as introns and transposons→ this in turn contributes to genome size without actually increasing the number of genes or biological complexity.
Genome
Total genetic information carried by all chromosomes of a cell organism.
Chromosome
Long threadlike structure composed of DNA and proteins that carry the genetic info of an organism
Gene
Segment of DNA that directs the formation of a protein or RNA. Unit of Heredity
E Coli
Bacterium single 4.6 Million Base Pair single circular chromosome
Humans have ____ chromosomes + ______
22 chromosomes + sex chromosomes (XX or XY)
Down Syndrome
Trisomy of chromosome 21 that causes developmental delays, intellectual disability and characteristic physical features.
Why is the older age of the mother correlated to Down syndrome?
Down syndrome is usually caused by a nondisjunction error, where chromosome 21 fails to separate properly during meiosis in the mother’s egg cell. As a woman ages the structure that hold chromosomes (like cohesion proteins and the spindle apparatus) weaken.
Are gene number and genome size correlated?
No much of an organism’s genome does not code for proteins. Large genomes often contain vast amounts of repetitive DNA, introns, and transposable elements, which increase genome size without adding more genes.
Closely Related Species can
have vastly different chromosome numbers
28% of the human genome contains genes yet only 1.5% of the genome is protein coding. What element explains this difference?
Although about 28% of the human genome is part of genes, most of that consists of introns (noncoding sequence) and untranslated regions (UTRs) that do not code for proteins. Only the exons, which make up roughly 1.5% of the genome are actually translated into amino acids.
What is most of intergenic DNA composed of?
Repetitive DNA sequences
Transposons
50% of the human genome consists of transposons-DNA elements that can move throughout the genome.
Transposons can copy or cut themselves out of one part of the genome and insert themselves into another. This movement can change how genes are expressed or even disrupt genes.
What are the effects of transposons?
By inserting into new places, transposons can create mutations, alter gene regulation and contribute to genetic diversity over time.
LINE
Long interspersed nuclear element
(retrotransposon-many kb in length)
SINE
Short interspersed nuclear element
(Transposable element 100-700 bp in length)
LTR Transposons
Long terminal repeat transposon (transposon that uses RNA as an intermediate)
Exons
Protein coding portion of gene
Intron
noncoding portion of genes. Must be removed by RNA splicing before RNA is translated
Which is longer, introns or exons?
Introns tend to be longer than exons.
How does DNA fit inside a nucleus?
DNA must be highly condensed to fit inside the nucleus
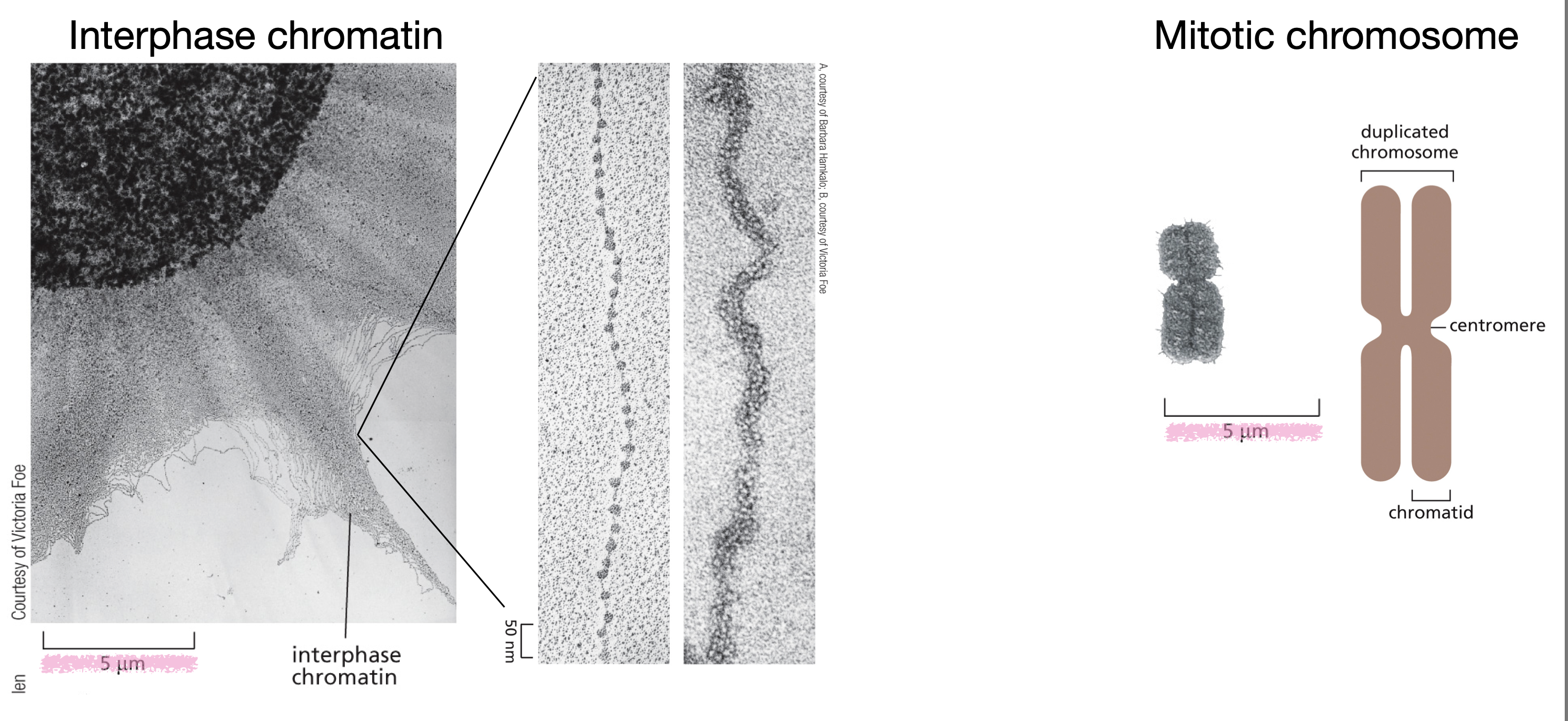
What is the difference between interphase chromosomes and mitotic chromosomes?
Interphase chromosomes are in the form of chromatin (uncoiled), are transcriptionally active and participate in DNA replication, and located inside the nucleus
Mitotic chromosomes are condensed into distinct, visible chromosomes, not transcriptionally active, consist of two sister chromatids attached to a centromere, and are located in the cytoplasm (spindle area) after nuclear envelope breaks down.
Chromatin
-The material of chromosomes (both protein and DNA)
-50% DNA and 50% protein
-Chromatin proteins are histones, structural proteins, DNA binding proteins, and transcription factors.
-The most basic unit of chromatin is the nucleosome which is DNA wrapped around a histone octamer
How does DNA packing occur at multiple levels?
First DNA is wrapped around nucleosomes like string around beads. Then these nucleosomes associate with each other to make a CHROMATIN fiber which are then looped around each other to build the entire mitotic chromosome.
What is the result of DNA packing through chromosomes?
Each DNA molecule has been packaged into a mitotic chromosome that is 10,000 fold shorter than its fully extended length.
Why are nucleosomes octamers?
They contain two copies of each of Histone 2A, Histone 2B, Histone 3, and Histone 4
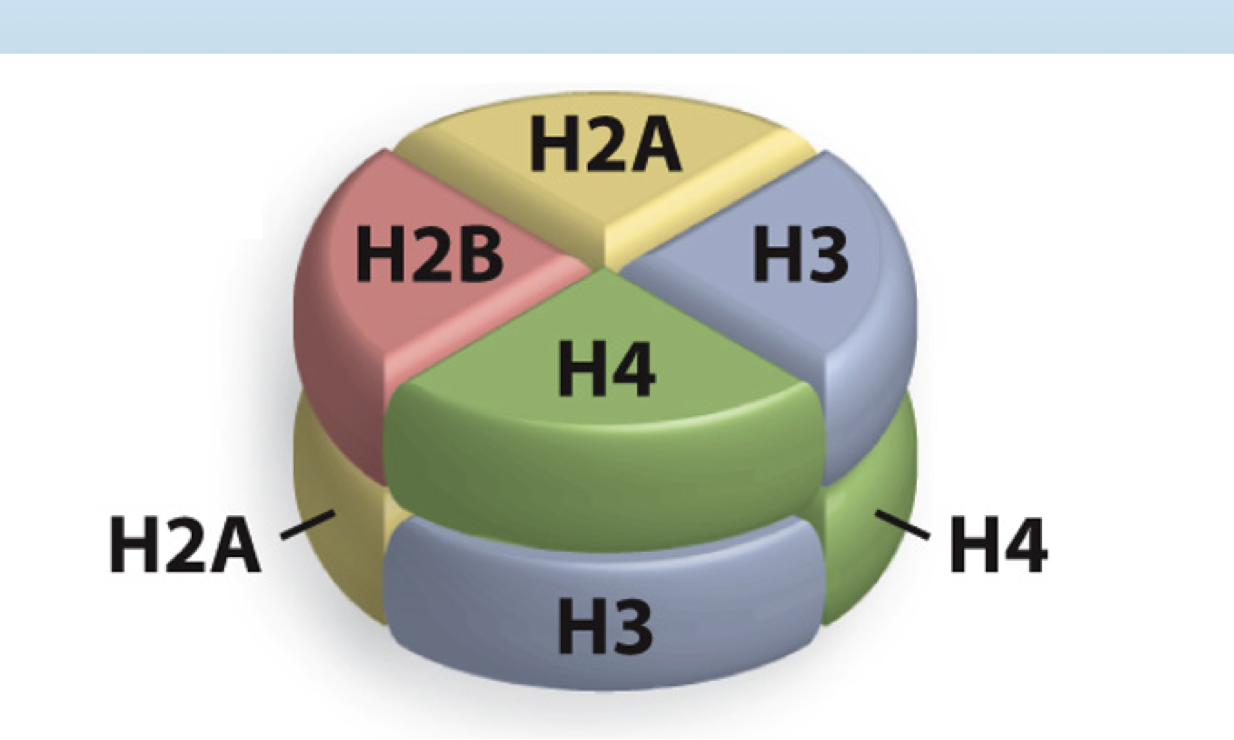
How much DNA is wrapped around the octamer of histones (nucleosome)?
147 base pairs
DNA forms a _________ around histones
left handed helical wrap
Histone H1
Linker Histone that has two DNA binding domains. H1 alters DNA entry by having enter and exit angles. This results in a 35-40 fold reduction in DNA length
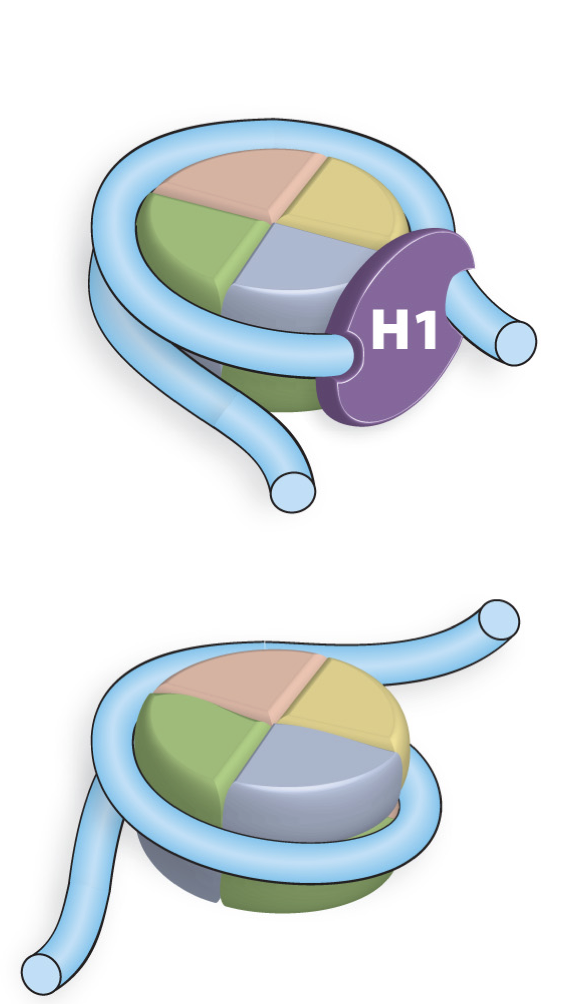
Composition of Histones
Small Highly basic proteins (K+R residues). These positive residues react favorably with the negatively charged phosphate backbone of DNA
Constituents of the histone fold
Three alpha helices that are connected by two short loops. This helix-loop-helix-loop pattern forms a handshake structure that allows histone proteins to pair up tightly with one another.
N termini modification of histones
The N termini of the Histone proteins extends beyond the nucleosome core and can be covalently modified by phosphorylation, acetylation, methylation, or ubiquitination
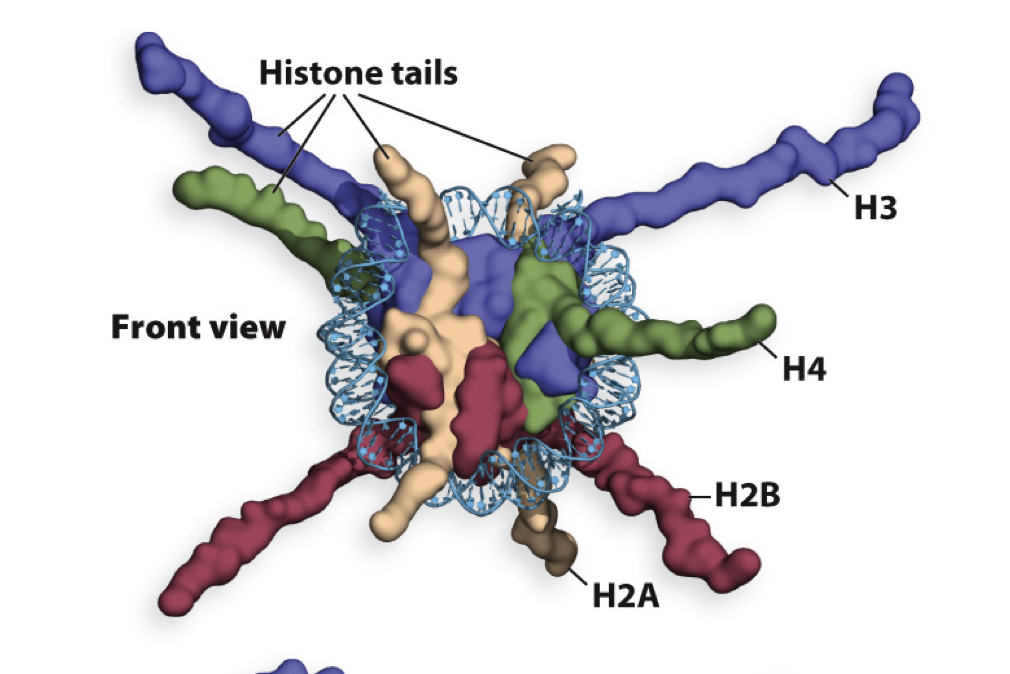
What does modification of N terminal tails of Histones do?
Changes the chemical properties of histones and interactions between nucelosomes and important for chromatin structure and function
What does modification of histone tails not do?
Modification of histone tails does not affect the interaction between histones and DNA