Gene expression
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
23 Terms
What are transcription regulatory proteins?
recognize specific DNA sequences because of the surface features of the double helix (recognizes surface features)
Complex arrangements that respond to a variety of signals that are interpreted and integrated
Double helix features = major & minor groove
Binds to the major grooves because it has the cis-sequences & other contact points
Contain structural motifs
Don't need to unwind DNA to read sequences
In bacteria: transcription is controlled by 1 regulatory protein (and have operons)
In eukaryotes: many proteins control transcription & can act over very long distances
Single transcription regulatory proteins
Single proteins can make the final decision as to transcribe or not
Can form an entire organ by beginning a cascade of other proteins & cell-cell interactions
Ex: eyeless drosophila (eyes instead of legs form)
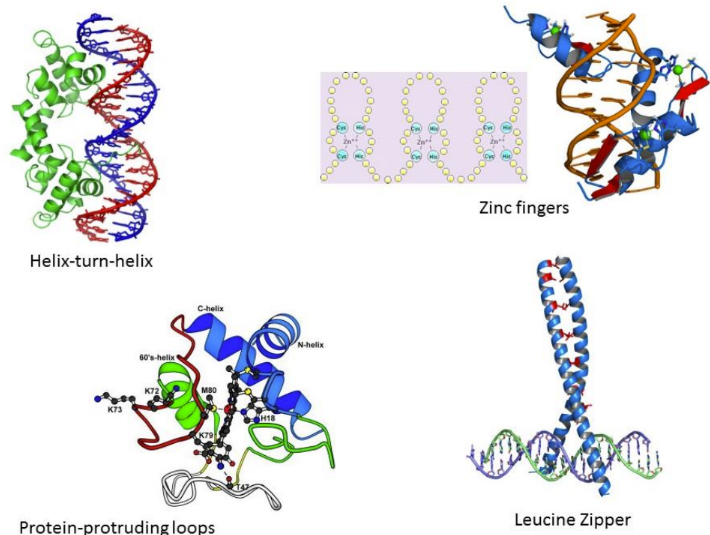
What are structural motifs
bind to the major groove based on the protein’s amino acid side chains (a recognizable, recurring 3D arrangement of secondary structure elements, such as helices and sheets, connected by loops)
Found in transcription regulators
Common motifs:
Helix-turn-helix = 2 alpha helices with a recognition sequence
Homeodomains are a type of helix-turn-helix
Zinc fingers = DNA binding proteins with a zinc (unique) moiety
zinc can hold alpha helix and beta sheet together
Zinc fills a two-alpha helix protein
Protein protruding loops =
Leucine zippers =
What are cis-regulatory sequence
DNA sequences specify the time and place that each gene is to be transcribed when read by their specific bound transcription regulators
DNA sequences recognized within the same molecule
Control transcription and are upstream of the transcription initiation start point
Positive regulators = activators
Negative regulators = repressors
Many protein regulators control a single gene (many regulator sequences within that gene
DNA looping critical for proteins, sequences, and polymerase to interact
“Complex switches”
What is the significance of DNA looping
DNA looping critical for complex switches to interact (it ISN’T LINEAR)
Brings activators/repressors and sequences closer to the mediator and other TFs
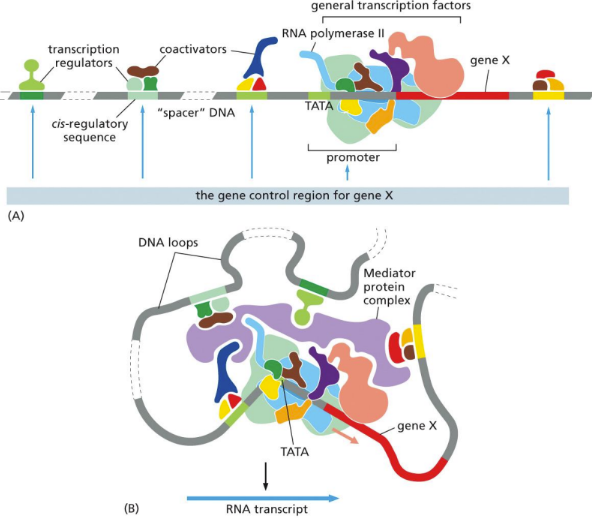
Gene control region (GCR)
consists of activator (and co-activator)/regulatory sequence/associated proteins, promoter region/factors/RNA pol, gene, more reg. proteins (chromatin remodelers)
Formed when a mediator is present and binds all the components together
RNA pol. II then transcribes all coding genes & non-coding RNA genes
How do cells control their own proteins? (list and describe)
Transcriptional control = DNA to RNA transcript
Controlled by DNA sequences (cis-regulatory sequences) near a gene upstream of the transcription initiation start point which are recognized by transcription regulator proteins
RNA processing
RNA transcript to mRNA
RNA transport and localization
mRNA being exported from the nucleus to the cytosol
Translation control
mRNA to protein
Degradation control (both mRNA & protein)
mRNA to inactive mRNA (degradation) or protein degraded into inactive protein
Protein activity control (phosphorylation)
Making a protein into active or inactive
How are cells specialized
Differences in RNA synthesis, transcription processing, and protein accumulation makes cells different and NOT DNA sequences
Large array of proteins control expression of certain genes (activators/repressors), that act at different times during cell development
Different combinations of proteins actions begin transcription of different genes
Controlled by an intracellular “gene control network” that responds to the environment (epigenetics)
Single transcription regulatory proteins can control transcription (more details on transcription regulatory proteins)
What do specialized cells have
Specialized cells:
Similarities in fundamental proteins (DNA/RNA polymerase, DNA repair enzymes, metabolic enzymes, etc)
Some proteins only made in the cells they function in
Levels of expression of active genes varies between cells
Alternative splicing and post-translational modifications occur in ALL cells
De-differentiating cells = manipulation of certain master transcription regulators (Oct4, Sox2, & Klf4) causing the cell to de-differentiate to become stem cells
The 3 TR are artificially expressed in fibroblasts -> lose specialization -> become pluripotent stem cells (used to treat diseases)
What are complex switches
control gene transcription in eukaryotes
involve regulatory elements like enhancers and promoters, often located in non-coding regions, that control gene expression
DNA looping critical for them to interact (it ISN’T LINEAR)
Longer stretches of DNA looped out and spaced out
More signals on one promoter
More factors & proteins
More developed in eukaryotes
What is a promoter
Nucleotide sequence in DNA to which RNA polymerase binds to begin transcription
Stepwise assembly of many transcription factors at promoter site can change rate of initiation
What is an enhancer
Sequences regulatory proteins (transcriptional activators) must bind to in DNA (called enhancers) to help attract the general transcription factors and RNA polymerase II to the start point of transcription
What is a mediator
30 subunit protein complex that acts as a bridge between all components of GCR (gene control region)
When present and binding everything, then it forms the GCR
What are activator proteins
proteins that bind cis-regulatory sequences (formerly enhancer sequences)
Can act at different steps during transcription initiation to promote it
Binding to RNA pol. To continue transcription by loading elongation factors
Releasing RNA pol. From promoter to start transcription
Bring RNA pol. To sequence
Can be far far away from the gene
Function = to attract, position, and modify transcription factors to get close to RNA pol
Helps recruit proteins, not only getting things started
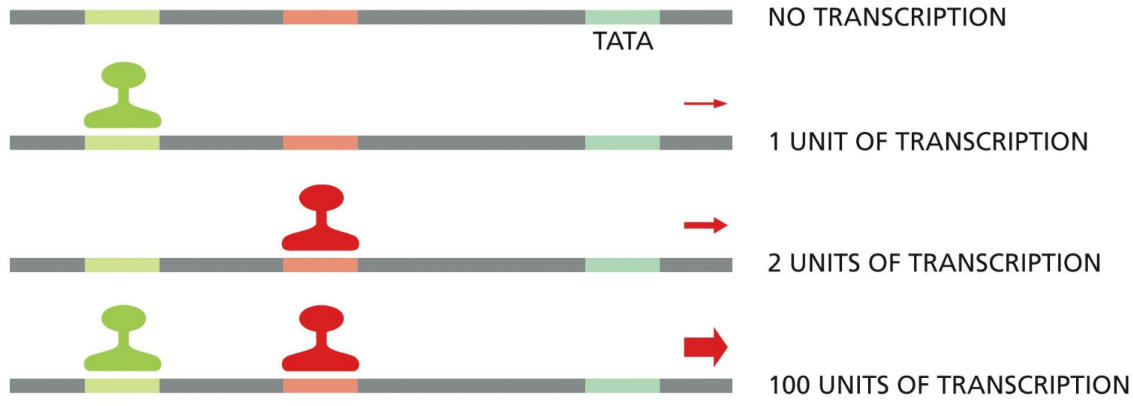
Can work synergistically to increase rate of transcription (works together)
transcriptional synergy
DNA looping brings activator and sequence closer to the mediator and other TF
What is transcriptional synergy
where several DNA-bound activator proteins working together produce a transcription rate that is much higher than the sum of their transcription rates working alone
What are repressor proteins
Proteins that further regulate transcription by using several mechanisms to ensure repression of transcription
Do NOT compete with RNA polymerase
Developmental regulatory genes are repressed after normal embryonic development
Sometimes they’re re-expressed with tumors (oncofetal antigens)
Ex: CEA (colon cancer type & alpha-fetoprotein in liver cancer
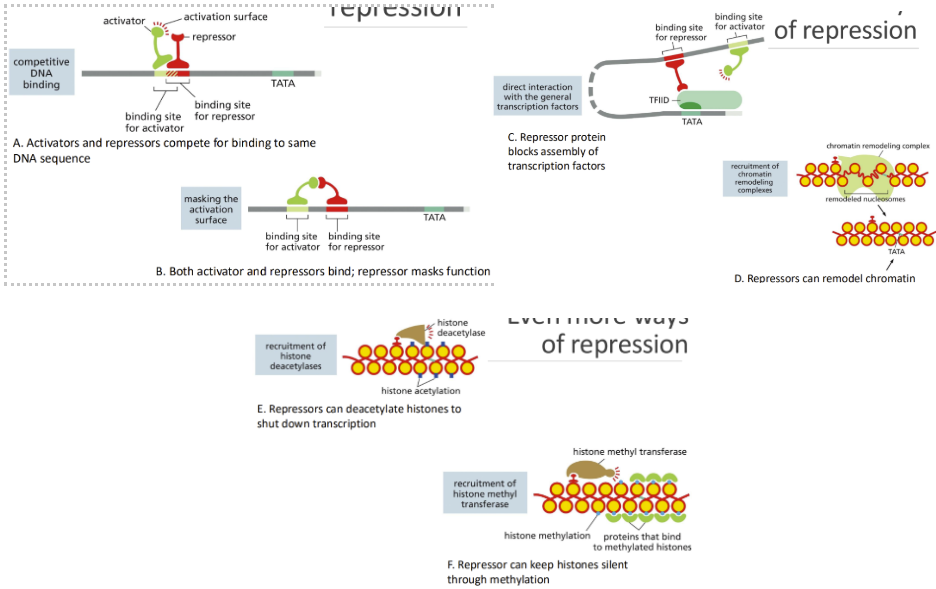
What are some ways repressions can occur
Ways of repression:
Competitive DNA binding = repressor competes with activator
Masking the activation surface = activator/repressor are close enough to cover the other’s domain
Direct interaction with the general transcription factors = pushes activator away, preventing it from interacting
Recruitment of chromatin remodeling complexes = remodel chromatin to prevent binding
Recruitment of histone deacetylases = shutdown transcription by deacetylating
Recruitment of histone methyl transferase = methylates histones to silence transcription
What are cis-regulatory sequences
Gene regulatory proteins MUST BE BOUND to DNA (either directly or indirectly) at the cis-regulatory sequence to influence transcription of a target gene
non-coding region that regulate transcription (where regulatory proteins bind)
Ex: enhancer
regulatory proteins = Activator or repressor
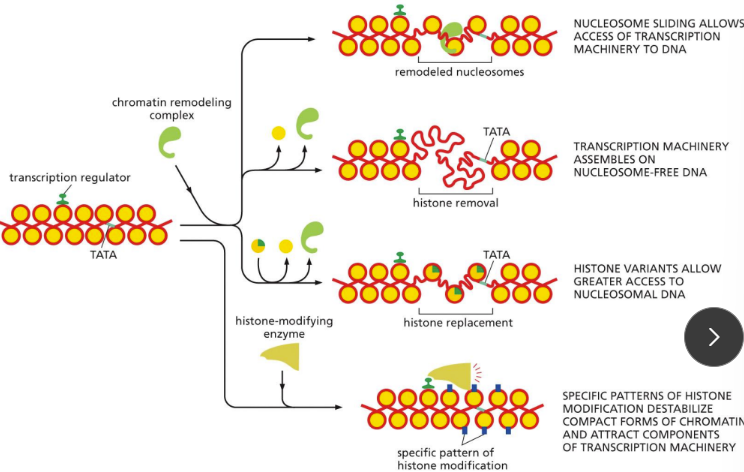
What are ways of chromatin/histone remodeling processes
Altering chromatin structure:
Activators can also alter chromatin structure to promote transcription
Makes underlying DNA more accessible to TFs, mediator, and RNA pol.
Needs to be accessed for transcription to happen
Mutant proteins block activators
Histone
Mutant proteins block transcription of histones
Possible ways:
Nucleosome sliding allows access of transcription machinery to DNA
Refold and open up
Transcription machinery assembles on nucleosome-free DNA
Unwinding and disassemble nucleosome (removing the nucleosomes)
Histone variants allow greater access to nucleosomal DNA
With histone chaperons
replacing the normal histones, with varients that allow for greater access
Specific patterns of histone modification destabilize compact forms of chromatin and attract components of transcription machinery
Via histone modifications
Why does X-inactivation occur
There is a dosage compensation in females (sometimes XXY males) because males only have 1X while females have 2X (one must get shut off)
Mutations that interfere are lethal
What is transcription inactivation
X chromosome in somatic cells are dosaged compensated by X-inactivation
Done to achieve dosage compensation (reduce X expression in females)
Maintained through replication and cell division
Early in cell development, one X in females is randomly condensed into heterochromatin (via Xist) and never recovers (not necessarily organ specific)
Some cells will express the maternal X and others the paternal X
How does transcription inactivation occur?
XIC is found in the middle of the X chromosome (X-inactivation center)
Both X chromosomes will have an XIC (only one will express Xist RNA = barr body)
Xist RNA will ONLY be expressed by the inactive X (not as a protein) and coat the chromosome -> driving it into a heterochromatin formation
15-20% of genes at the tips of the chromatin loops remain active
Histone variant with many modifications makes transcription of the inactive X virtually impossible
How are females mosaic
Because of randomly inactivated Xs
Reversed during germ cell formation, so haploid germ cells have an active X
What is oncofetal antigens
Oncofetal antigens = re-expressed developmental regulatory genes, after normal embryonic development, resulting in tumors
Ex: Carcinoembryonic antigen (CEA) in certain colon cancers & alpha-fetoprotein in liver cancer